Pediatric Therapeutics and Pharmacology
Session: Pediatric Therapeutics and Pharmacology
817 - Pharmacogenomic Implications of Genes on the American College of Medical Genetics Secondary Findings List
Friday, April 25, 2025
5:30pm - 7:45pm HST
Publication Number: 817.6355
Benjamin Q. Duong, Nemours Children's Hospital, Boca Raton, FL, United States; Josiah Allen, St. Elizabeth Healthcare, Edgewood, KY, United States; Jordan Brady, St. Elizabeth, Edgewood, OH, United States; Pamela Arn, Nemours Children's Health, Jacksonville, FL, United States

Benjamin Q. Duong, PharmD (he/him/his)
Clinical Pharmacogenomics Service Manager
Nemours Children's Health
Boca Raton, Florida, United States
Presenting Author(s)
Background: Historically, the clinical genomics (CGx) and pharmacogenomics (PGx) domains have remained largely distinct. CGx assesses genetic variants associated with disease diagnosis and risk while PGx evaluates genes associated with drug metabolisms and response. However, we have identified multiple cases that demonstrate the intersection between CGx and PGx variants. In one case, a child with a pathogenic variant SDHA, associated with risk of pheochromocytoma, was prescribed atomoxetine for ADHD which may interfere with the annual screenings of metanephrine levels.
Objective: The aim of this study is to explore the overlap in genes with both disease associations and PGx implications within the American College of Medical Genetics Secondary Findings list v3.2 (ACMG SF v3.2) of recommended genes.
Design/Methods: We reviewed the ACMG-SF v3.2 genes with two common genomic knowledgebases: GeneReviews and PharmGKB. Targeted therapies for specific germline/somatic variants were excluded. Interactions were rated on a 1-4 scale by pharmacogenomics-trained pharmacists and a genetic counselor: Level 1 indicates FDA or guideline-driven recommendations, Level 2 indicates clinically significant interactions, Level 3 indicates potential interaction but no recommendations, and Level 4 indicates no/weak interaction. The FDA drug product label database was then queried if any drug’s “Contraindications” and “Warnings and Precautions” section referenced a Level 1 or 2 disease implication.
Results: Of the 81 genes reviewed, 30 (37%) had Level 1 or Level 2 interactions. PharmGKB identified 3/4 (75%) Level 1 interactions and 5/26 (19%) Level 2 interactions. GeneReviews identified 3/4 (75%) Level 1 interactions and 19/26 (73%) Level 2 interactions. The FDA Label Database had contraindications or warnings labeling specific to 4/4 (100%) Level 1 interactions and 7/26 (27%) Level 2 interactions. Examples include risks inferred by the disease pathology (e.g. stimulants and catecholaminergic polymorphic ventricular tachycardia) and less obvious associations (e.g. Marfan syndrome and fluoroquinolones).
Conclusion(s): Our results emphasize a need for greater research on medication implications of hereditary disease genes. There is concern for patient harm if these interactions are not known, assessed, and communicated appropriately via education and electronic clinical decision support. The limited literature and drug labeling on this topic makes risk identification/triage challenging. As clinical practice continues to move towards broad preemptive genetic testing, practitioners must be aware of both CGx and PGx implications of genetic findings.
Figure 1. Scale of Pharmacogenomic Implications
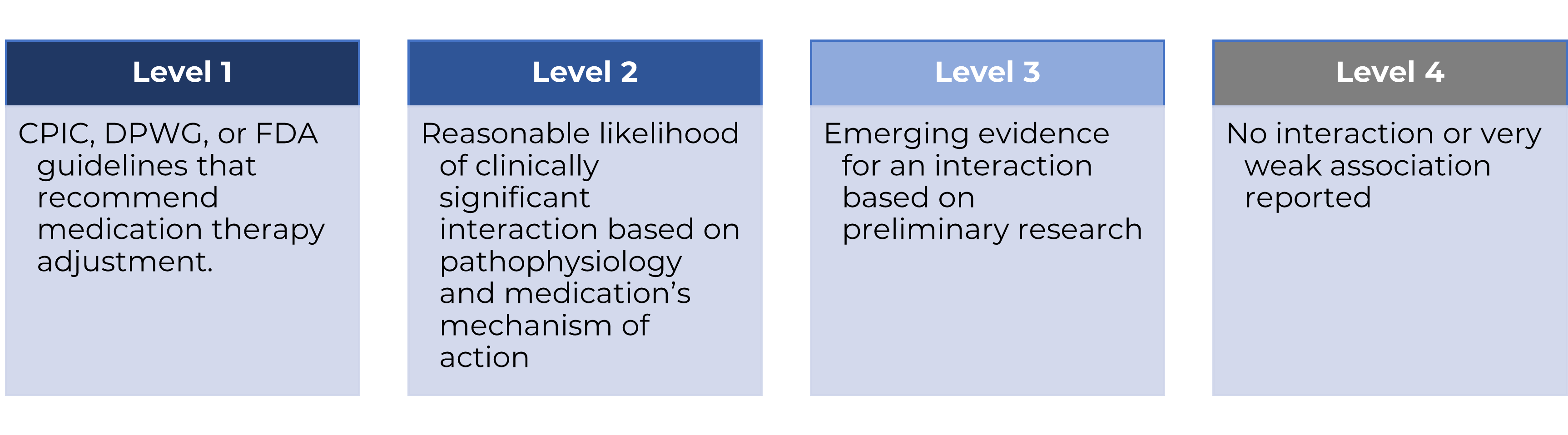
Figure 2. ACMG Genes by Interaction Level and Source
.png)
Table 1: Level 1 and Level 2 Genes with Clinical Genomics and Pharmacogenomics Implications
.png)

