Neonatal Pulmonology - Basic/Translational Science 2
Session: Neonatal Pulmonology - Basic/Translational Science 2
220 - Commensal Oral Microbiota Associated With Bronchopulmonary Dysplasia Development
Friday, April 25, 2025
5:30pm - 7:45pm HST
Publication Number: 220.4216
Samuel Gentle, Yale School of Medicine, Guilford, CT, United States; Karina Ricart, University of Alabama School of Medicine, Birmingham, AL, United States; Nengjun Yi, University of Alabama at Birmingham, Birmingham, AL, United States; Kent A.. Willis, The University of Alabama at Birmingham, Birmingham, AL, United States; Charitharth V. Lal, University of Alabama School of Medicine, Birmingham, AL, United States; Namasivayam Ambalavanan, University of Alabama School of Medicine, Birmingham, AL, United States; Rakesh P. Patel, University of Alabama School of Medicine, Birmingham, AL, United States

Samuel Gentle, MD (he/him/his)
Assistant Professor
Yale School of Medicine
Guilford, Connecticut, United States
Presenting Author(s)
Background: Airway dysbiosis has been implicated in the development of bronchopulmonary dysplasia (BPD). However, the diagnostic potential for dysbiosis in the preterm oral microbiome in the context of BPD development has been understudied.
Objective: We hypothesized that buccal microbial diversity and individual taxa would differ between infants with and without BPD development and within infants with BPD by lung severity.
Design/Methods: Single center prospective cohort study in infants born at < 29 weeks’ gestation between 2020 and 2022. Samples from the tongue dorsum were collected at four postnatal time points: birth, 27-, 29-, and 36 weeks’ postmenstrual age. 16s rRNA sequencing analyses were performed at: 1) the microbial community level to relate overall patterns of taxonomic composition to comparison groups and 2) the single-taxon level to identify individual taxa significantly associated lung disease. For the community-level analyses, alpha and beta diversities were used as the overall microbial measures, and test associations between diversities and comparison groups of interest determined. A negative binomial additive model was performed for the single-taxon analysis by treating the microbiome counts as response and group indicator and splines of time as predictors. Comparison groups included 1) BPD vs no BPD, 2) BPD associated pulmonary hypertension (BPD-PH) vs no BPD-PH, and 3) Grade 2-3 BPD vs no Grade 2-3 BPD per Jensen criteria.
Results: Data from 64 infants were available for analysis of whom 40 developed BPD, 16 did not develop BPD, and 8 died prior to 36 weeks’ PMA (Table 1). Alpha (Figure 1) and beta diversity did not differ between any of the evaluated comparison groups. In longitudinal analyses of single-taxon level differences, Staphylocuccus aureus, Actinomyces, and Corynebacterium differed over time between infants with and without BPD. Bacteroides was less commonly identified in infants that developed pulmonary hypertension. Lastly, Enterobacter, Micrococcus, and Peptoniphilus differed by BPD severity (Figure 2).
Conclusion(s): While buccal diversity indices did not differ by comparison group, microbial signatures differed within all comparison groups with potential predictive and therapeutic implications.
Table 1: Demographic and Clinical Characteristics by Comparison Groups
Table 1.pdf1median (interquartile range)
*p value for comparisons between infants with No BPD and BPD
Alpha Diversity Analyses
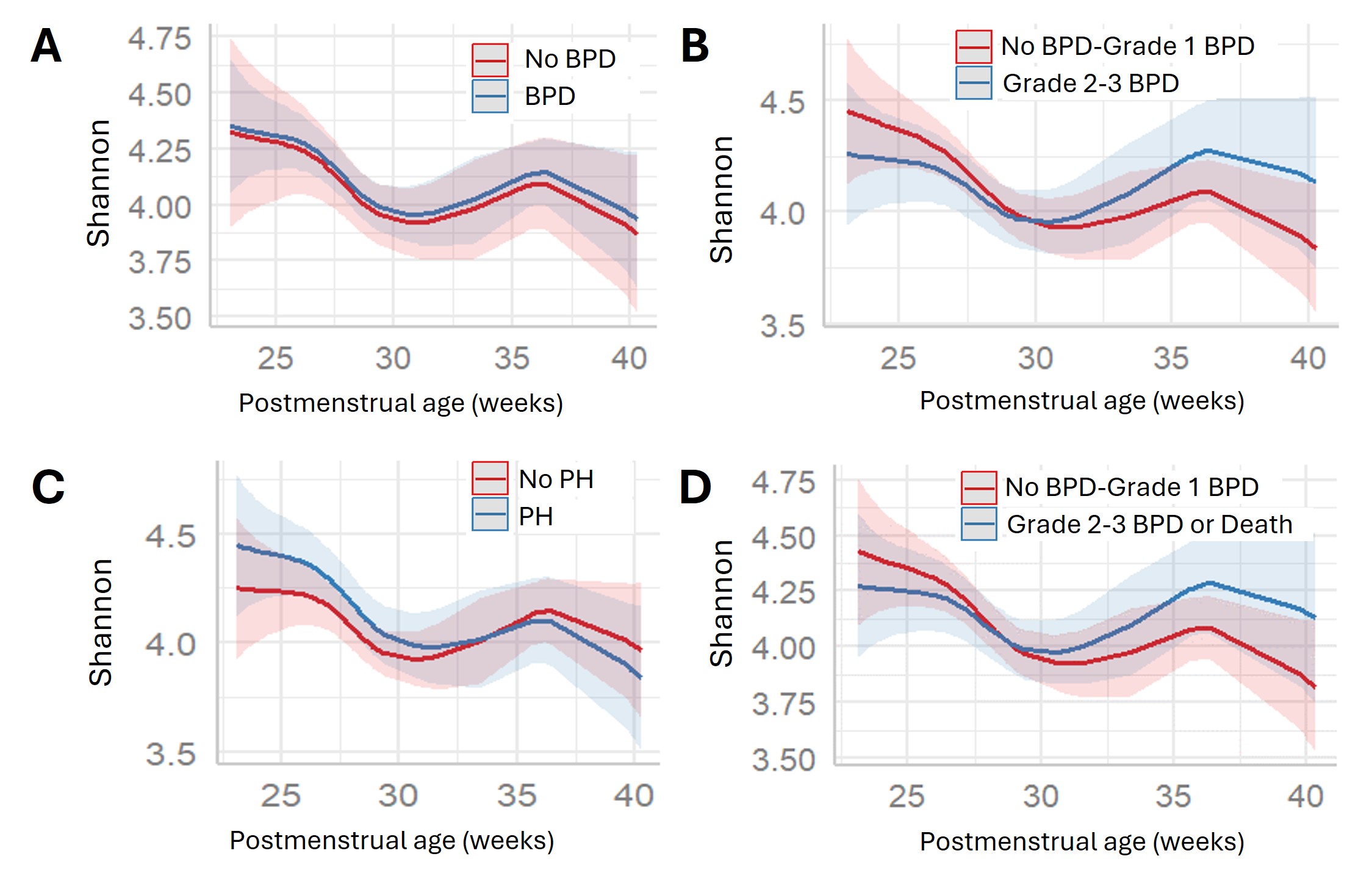 Alpha diversity plots over time between comparison groups of A: No BPD vs BPD, B: No BPD/Grade 1 BPD vs Grade 2-3 BPD, C: No pulmonary hypertension vs Pulmonary Hypertension, and D: No BPD/Grade 1 BPD vs Grade 2-3 BPD or death. No measures of alpha diversity significantly differed by comparison groups (A: p=0.96, B: p=0.07, C: p=0.22, D: p=0.09).
Alpha diversity plots over time between comparison groups of A: No BPD vs BPD, B: No BPD/Grade 1 BPD vs Grade 2-3 BPD, C: No pulmonary hypertension vs Pulmonary Hypertension, and D: No BPD/Grade 1 BPD vs Grade 2-3 BPD or death. No measures of alpha diversity significantly differed by comparison groups (A: p=0.96, B: p=0.07, C: p=0.22, D: p=0.09).Single-Taxon Analyses
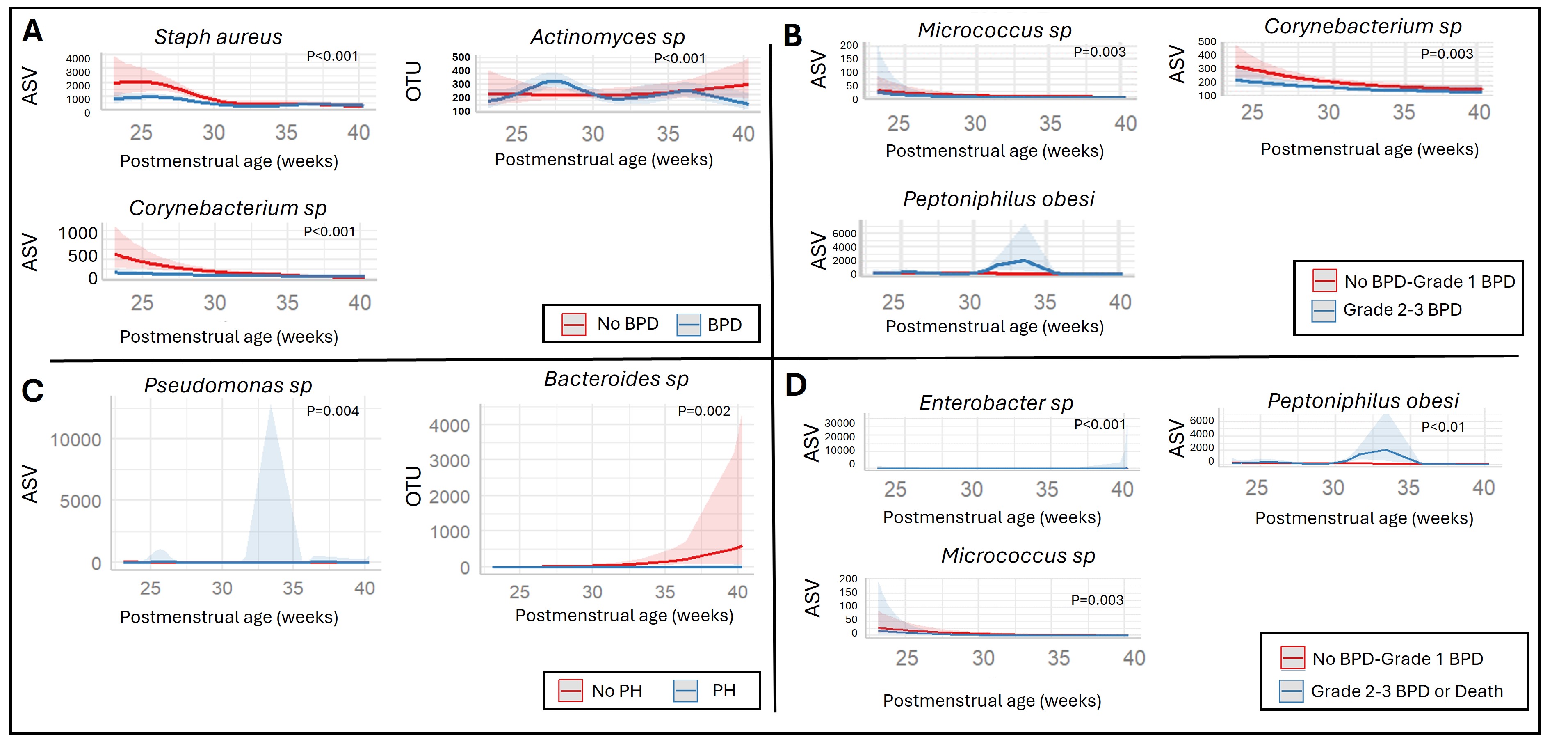 Single-taxon significant differences over time between comparison groups of A: No BPD vs BPD, B: No BPD/Grade 1 BPD vs Grade 2-3 BPD, C: No pulmonary hypertension vs Pulmonary Hypertension, and D: No BPD/Grade 1 BPD vs Grade 2-3 BPD or death.
Single-taxon significant differences over time between comparison groups of A: No BPD vs BPD, B: No BPD/Grade 1 BPD vs Grade 2-3 BPD, C: No pulmonary hypertension vs Pulmonary Hypertension, and D: No BPD/Grade 1 BPD vs Grade 2-3 BPD or death. 
