Genomics/Epigenomics 2
Session: Genomics/Epigenomics 2
399 - Investigating diagnostic, prognostic, and therapeutic gene targets in hepatoblastoma
Friday, April 25, 2025
5:30pm - 7:45pm HST
Publication Number: 399.5139
Sarah Voskamp, University of Central Florida College of Medicine, Orlando, FL, United States; Jennifer S. Nelson, University of Central Florida College of Medicine, Orlando, FL, United States

Sarah Voskamp, BS (she/her/hers)
Medical Student
University of Central Florida College of Medicine
Orlando, Florida, United States
Presenting Author(s)
Background: Hepatoblastoma is the most common malignant pediatric liver cancer, and the incidence has been rising. Despite numerous advancements in the understanding of hepatoblastoma and the genetic and epigenetic influences, a definitive etiology has yet to be identified.
Objective: Herein we present an investigation of the genetic signatures associated with hepatoblastoma, highlighting additional genes that may prove to be causative mutations, potential therapeutic targets, or diagnostic biomarkers.
Design/Methods: The Search Tag Analyze Resource for NCBI’s Gene Expression Omnibus (STARGEO) was utilized to identify 138 hepatoblastoma tumor samples and 41 healthy liver samples for investigation. Meta-analysis was performed and associated gene signatures were analyzed using Ingenuity Pathway Analysis (IPA) and restricted to genes with a statistically significant difference (p < 0.05) and absolute experimental log ratio greater than 0.2 between hepatoblastoma and healthy liver.
Results: 2203 molecules were identified as having significant differential expression between hepatoblastoma and healthy liver. Top upregulated genes include MAGED4/MAGED4B, SLC711A, WIF1, CENPA, and DKK1 and are largely involved in transcription regulation and inhibition of the beta-catenin/Wnt signaling pathway. The top canonical pathways include Selenoamino acid metabolism, phase 1 - functionalization of compounds, and cell cycle checkpoints. The top upstream regulators are dexamethasone, CTNNB1, beta-estradiol, HNF4A, and CEBPB. Toxicity functions associated with hepatoblastoma include growth of hepatocellular carcinoma and proliferation of liver cancer cells.
Conclusion(s): Identifying genetic signatures associated with hepatoblastoma is a necessary preliminary step to develop efficacious therapeutic and diagnostic strategies. Potential prognostic biomarkers include MAGED4/MAGED4B and DKK1 as upregulation of both genes is associated with poor survival. HMGA2 is a diagnostic marker useful in distinguishing hepatoblastoma from hepatocellular carcinoma. Therapeutic strategies warranting further investigation include knockdown of SLC7A11, silencing of CENPA, and utilization of the novel cell-cycle inhibitor JNJ-7706621.
Figure 1. Top canonical pathways
.jpg) Top canonical pathways demonstrating differential expression in hepatoblastoma. Larger bubbles depict a greater amount of differentially expressed genes within the pathway.
Top canonical pathways demonstrating differential expression in hepatoblastoma. Larger bubbles depict a greater amount of differentially expressed genes within the pathway.Figure 2. Proliferation of liver cancer cells
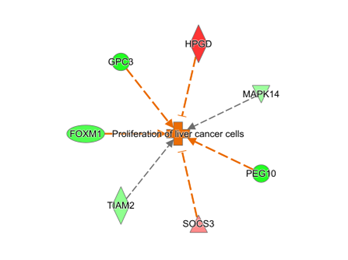 Network associated with top toxicity function, proliferation of liver cancer cells.
Network associated with top toxicity function, proliferation of liver cancer cells.Figure 3. MAGED4 and DKK1 Survival
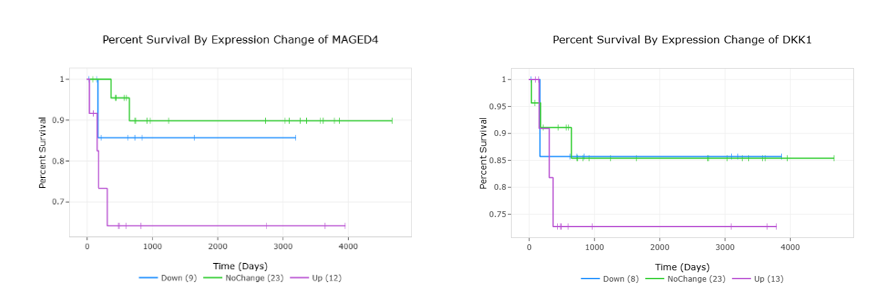 Percent survival stratified by expression of (a) MAGED4 and (b) DKK1 in hepatoblastoma samples.
Percent survival stratified by expression of (a) MAGED4 and (b) DKK1 in hepatoblastoma samples.Figure 1. Top canonical pathways
.jpg) Top canonical pathways demonstrating differential expression in hepatoblastoma. Larger bubbles depict a greater amount of differentially expressed genes within the pathway.
Top canonical pathways demonstrating differential expression in hepatoblastoma. Larger bubbles depict a greater amount of differentially expressed genes within the pathway.Figure 2. Proliferation of liver cancer cells
 Network associated with top toxicity function, proliferation of liver cancer cells.
Network associated with top toxicity function, proliferation of liver cancer cells.Figure 3. MAGED4 and DKK1 Survival
 Percent survival stratified by expression of (a) MAGED4 and (b) DKK1 in hepatoblastoma samples.
Percent survival stratified by expression of (a) MAGED4 and (b) DKK1 in hepatoblastoma samples.
