Infectious Diseases 3: Acute respiratory infections
Session: Infectious Diseases 3: Acute respiratory infections
130 - Comparison of multiplex RT-PCR and RNA-Seq for detection of pathogens in tracheal aspirate samples in critically ill children.
Saturday, April 26, 2025
2:30pm - 4:45pm HST
Publication Number: 130.4413
David I. Del Valle Ortiz, University of Rochester School of Medicine and Dentistry, San Juan, Puerto Rico, United States; Lauren Gunn-Sandell, University of Colorado, Aurora Medical Campus, Aurora, CO, United States; Padmini Deosthale-Bapat, University of California, San Francisco, School of Medicine, San Francisco, CA, United States; Emily Lydon, University of California, San Francisco, School of Medicine, San Francisco, MA, United States; Lilliam Ambroggio, Children's Hospital Colorado, Aurora, CO, United States; Brandie Wagner, University of Colorado School of Medicine, Aurora, CO, United States; Nadir Yehya, CHOP, PHILADELPHIA, PA, United States; Aline B. Maddux, University of Colorado School of Medicine, Aurora, CO, United States; Matthew Leroue, University of Colorado School of Medicine, Aurora, CO, United States; Eric A.F.. Simões, Children's Hospital Colorado, Aurora, CO, United States; Charles R. Langelier, UCSF, San Francisco, CA, United States; Samuel Dominguez, Children's Hospital Colorado, Denver, CO, United States; Peter M. Mourani, University of Arkansas for Medical Sciences, Little Rock, AR, United States; Christina M. Osborne, CHILDREN'S HOSPITAL OF PHILADELPHIA, Philadelphia, PA, United States

David I. Del Valle Ortiz (he/him/his)
Undergraduate Student
University of Rochester
San Juan, Puerto Rico, United States
Presenting Author(s)
Background: Bacterial lower respiratory tract infection (LRTI) is a significant cause of morbidity and mortality in children with who require mechanical ventilation (MV). Microbiologic diagnosis is made with culture or multiplex reverse transcriptase polymerase chain reaction (RT-PCR) from tracheal aspirates (TA), but given the limitations of each modality, a true gold standard test remains elusive. Unbiased metatranscriptomic sequencing (RNA-Seq) may enhance diagnosis but has not been evaluated against newer clinical RT-PCR tests that incorporate common bacterial pathogens.
Objective: Compare LRTI pathogen detection/nomination by RNA-Seq, RT-PCR, and culture on TAs from children requiring MV and suspected LRTI.
Design/Methods: Critically ill children (30 days-17 years) requiring MV via an endotracheal tube for >72 hours were enrolled prospectively from a tertiary care pediatric hospital. TAs were collected for the purposes of the study and were paired with clinical cultures sent for suspected LRTI obtained within 24 hours of the research sample. Research samples were analyzed by RT-PCR (Biofire FilmArray Pneumonia Panel) and RNA-Seq, and culture results were extracted from the medical record. Pathogens were identified from RNA-Seq data using a previously developed rules-based model based on dominance in the sample. Sensitivity, specificity, conditional logistic regression with pairwise comparison, and positive and negative percent agreement were used for comparison between tests.
Results: We compared 59 samples with results from culture, RT-PCR, and RNA-Seq. Pathogens detected by testing modality are shown in Figure 1A with 28/59 (47.5%) positive by all three modalities. Compared to culture, RNA-Seq had sensitivity of 65.2% (95% confidence interval [CI] 42.7%-83.6%) and specificity of 77.8% (95% CI 60.9%-89.9%) with similar likelihood of nominating pathogens (p = 0.96). Compared to RT-PCR, RNA-Seq had sensitivity of 82.6% (95% CI 61.2%-95.1%) and specificity of 36.1% (95% CI 20.8 -53.8%), and likelihood of nominating pathogens was significantly less (p < 0.001). Negative percent agreement was >90% across all organisms when comparing test modalities with variable positive percent agreement (Tables 1& 2). Pathogen nomination rates by sample and testing modality is shown in Figure 1B for the four most common organisms.
Conclusion(s): RNA-Seq of tracheal aspirate samples showed variable pathogen nomination rates when compared with culture and RT-PCR and had similar rates to culture. Given high detection rates by RT-PCR, it is possible that RT-PCR might be overly sensitive when used on samples from a non-sterile site.
Figure 1
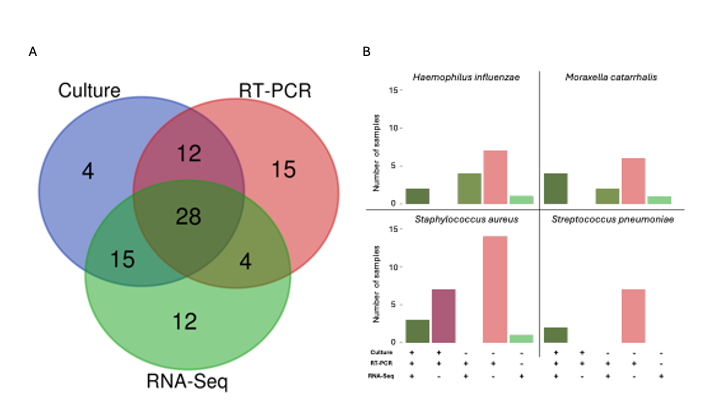 Figure 1- A) Number of pathogens nominated by each modality with overlap where pathogens were suggested by more than one modality. B) Rate of nomination by sample and testing modality for the four most common organisms (Haemophilus influenzae, Moraxella catarrhalis, Streptococcus pneumoniae, and Staphylococcus aureus).
Figure 1- A) Number of pathogens nominated by each modality with overlap where pathogens were suggested by more than one modality. B) Rate of nomination by sample and testing modality for the four most common organisms (Haemophilus influenzae, Moraxella catarrhalis, Streptococcus pneumoniae, and Staphylococcus aureus).Table 1
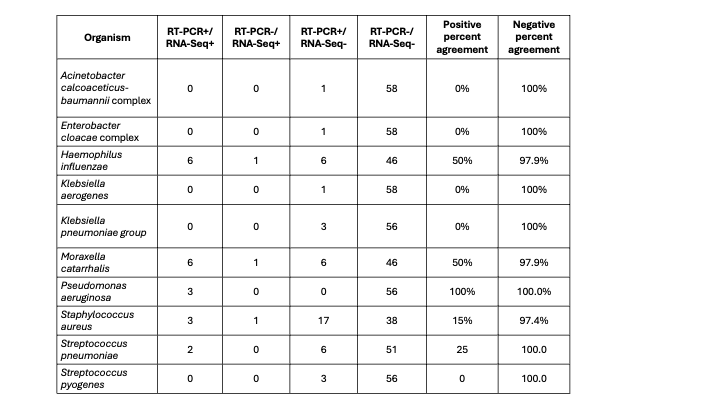 Table 1: Evaluation of pathogens by organism between RT-PCR and RNA-Seq. Positive percent agreement calculated as concordant positive/all positive results by RT-PCR
Table 1: Evaluation of pathogens by organism between RT-PCR and RNA-Seq. Positive percent agreement calculated as concordant positive/all positive results by RT-PCRTable 2
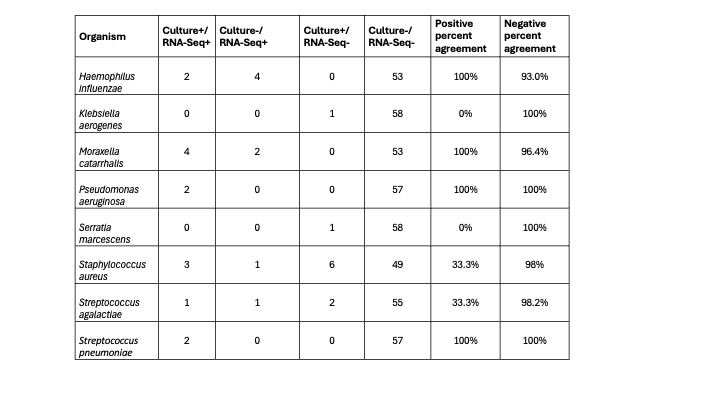 Table 2: Evaluation of pathogens by organism between culture and RNA-Seq. Positive percent agreement calculated as concordant positive/all positive results by culture
Table 2: Evaluation of pathogens by organism between culture and RNA-Seq. Positive percent agreement calculated as concordant positive/all positive results by culture
