Nephrology 3
Session: Nephrology 3
590 - The Clinical Utility of Next-Generation Sequencing-Based Genetic Testing in Pediatric Polycystic Kidney Disease
Sunday, April 27, 2025
8:30am - 10:45am HST
Publication Number: 590.6595
Elif G. Bozkurt, Mayo Clinic Children's Center, Rochester, MN, United States; Hana Yang, Mayo Clinic, Rochester, MN, United States; Tracy Baker, Mayo Clinic Children's Center, Rochester, MN, United States; Mohamad Sheikh Najeeb, Mayo Clinic Children's Center, Rochester, MN, United States; Whitney S. Thompson, Mayo Clinic, Rochester, MN, United States; Filippo Pinto e Vairo, Mayo Clinic, Rochester, MN, United States; Jennifer Kemppainen, Mayo Clinic Children's Center, Rochester, MN, United States; Cheryl Tran, Mayo Clinic Children's Center, Rochester, MN, United States; David J.. Sas, Mayo Clinic Children's Center, Rochester, MN, United States; Carl Cramer, Mayo Clinic Children's Center, Rochester, MN, United States; Fouad Chebib, Mayo Clinic Alix School of Medicine, Jacksonville, FL, United States; Neera Dahl, Mayo Clinic Alix School of Medicine, Rochester, MN, United States; Vicente Torres, Mayo Clinic, Rochester, MN, United States; Harris Peter, Mayo Clinic, Rochester, MN, United States; Christian Hanna, Mayo Clinic, Rochester, MN, United States
- EB
Elif G. Bozkurt, MD (she/her/hers)
Research Fellow
Mayo Clinic Children's Center
Rochester, Minnesota, United States
Presenting Author(s)
Background: Polycystic kidney disease (PKD) in children shows broad genetic heterogeneity such as ADPKD, ARPKD, and other ciliopathies, often making gene-specific testing inadequate and leading to missed diagnosis. Precise genetic identification in pediatric PKD is crucial for timely intervention and informed prognosis discussions, yet this remains challenging due to extensive genetic, allelic, and phenotypic variability.
Objective: To evaluate the effectiveness of next-generation sequencing (NGS)-based testing in diagnosing pediatric PKD, and to correlate genotype with phenotype.
Design/Methods: This prospective study, conducted from Jan 2020 to Jun 2024, included children under 18 with either two or more bilateral kidney cysts without a family history (FHx) of PKD, or at least one cyst with a positive FHx. Data were extracted from electronic medical records, and samples were collected from patients lacking clinical genetic testing or those with negative results that enabled research genetic testing, as well as, when possible, from family members.
Results: We evaluated 109 patients (53.2% female), with a median age of 9 years (range, 0-13), from 99 families. Genetic testing was completed for 93 patients (from 84 families), with 6 patients awaiting results, and 10 not providing samples. Initial testing identified pathogenic or likely pathogenic (P/LP) variants in 65/93 patients (69.9%) from 59/84 families (70.2%). The remaining 28 patients (25 families) were unresolved due to variants of uncertain significance (VUS) or benign variants. Segregation testing in relatives led to variant reclassification in 12/28 patients (9/25 families). Consequently, genetic etiology was identified in 77/93 patients (82.8%) from 68/84 families (80.9%). Variants were identified across a range of genes, including monoallelic or copy number changes in PKD1, PKD2, IFT140, NEK8, GANAB, PKHD1, TSC2, TSC2/PKD1, HNF1B, and ZMYM2, as well as biallelic variants in NPHP1 and TMEM67, and biallelic or suspected biallelic variants in PKHD1 associated with ARPKD phenotypes (Figures 1A, B). Intrauterine presentations (n=13) were linked to PKD1, HNF1B, 17q12 deletion, PKHD1 (biallelic), NEK8, and TSC2/PKD1. Monoallelic rare variants in IFT140, NEK8, GANAB, and PKHD1 (monoallelic) were found in children, alongside a novel ZMYM2 variant causing PKD. Figures 2 and 3 show NGS-based testing yield by presentation type, with resolved case percentages and distribution of causative genes.
Conclusion(s): NGS-based genetic testing showed high diagnostic yield and genetic diversity, underscoring its power in identifying disease-causing genes in pediatric PKD cases.
Fig 1. (A) Disease-causing genes and their frequency in genetically resolved patients (N=77). (B) Disease-causing genes and their frequency in genetically resolved families (N=68).
.png) Variants were identified across a range of genes, including monoallelic or copy number changes in PKD1, PKD2, IFT140, NEK8, GANAB, PKHD1, TSC2, TSC2/PKD1, HNF1B, and ZMYM2, as well as biallelic variants in NPHP1 and TMEM67, and biallelic or suspected biallelic variants in PKHD1 associated with ARPKD phenotypes.
Variants were identified across a range of genes, including monoallelic or copy number changes in PKD1, PKD2, IFT140, NEK8, GANAB, PKHD1, TSC2, TSC2/PKD1, HNF1B, and ZMYM2, as well as biallelic variants in NPHP1 and TMEM67, and biallelic or suspected biallelic variants in PKHD1 associated with ARPKD phenotypes. Fig 2. Genetic Diagnostic Yield by Presentation Type:
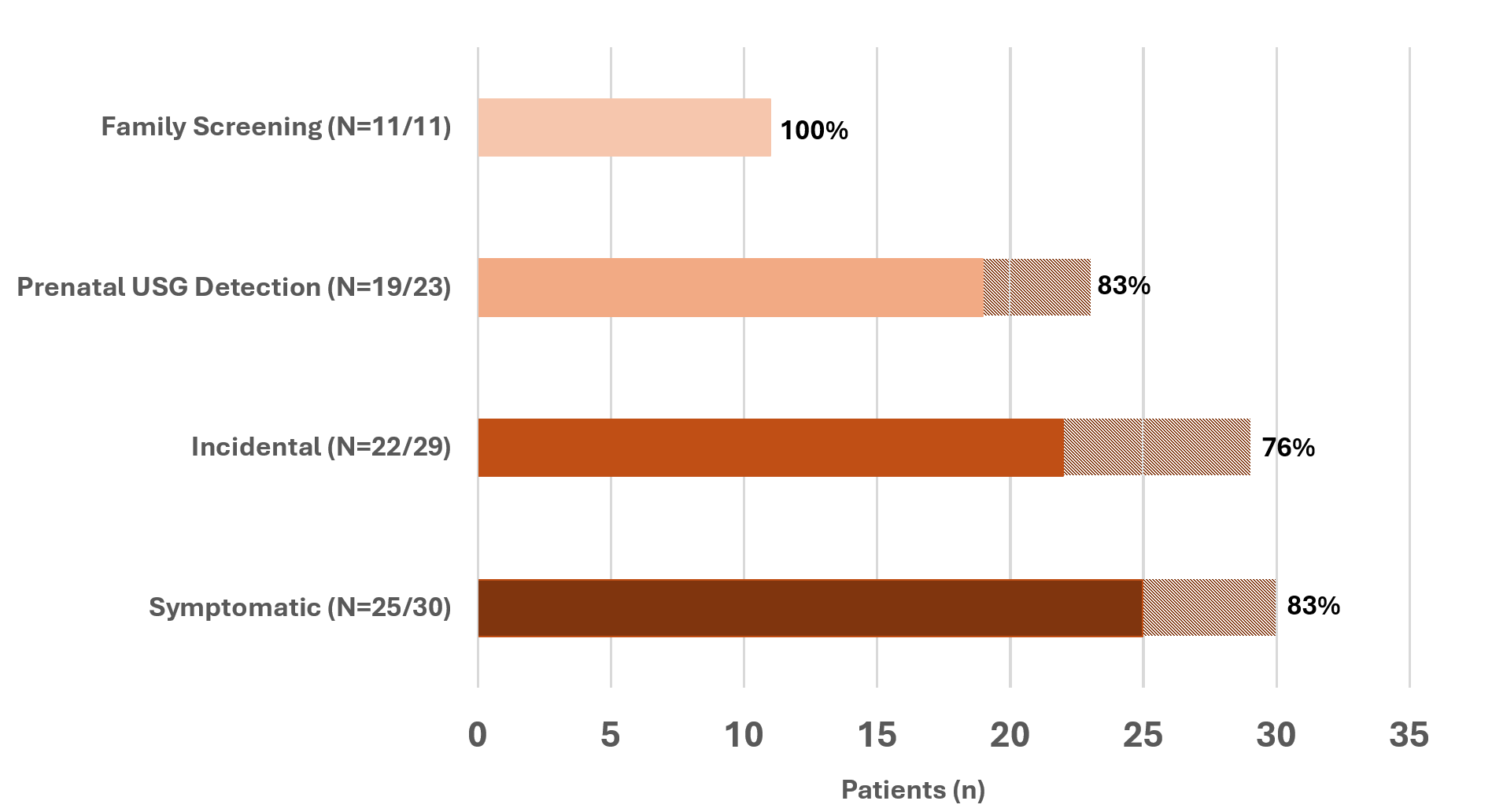 The y-axis categorizes initial presentation types: Family Screening, Prenatal Ultrasound (USG) Detection, Incidental, and Symptomatic. The x-axis shows the patient count. Solid-colored bars represent patients with resolved genetic diagnoses, with percentages indicating the resolution rate within each presentation type (Resolved patients, N=77). Dashed sections show the total number of patients tested per category (Tested patients, N=93).
The y-axis categorizes initial presentation types: Family Screening, Prenatal Ultrasound (USG) Detection, Incidental, and Symptomatic. The x-axis shows the patient count. Solid-colored bars represent patients with resolved genetic diagnoses, with percentages indicating the resolution rate within each presentation type (Resolved patients, N=77). Dashed sections show the total number of patients tested per category (Tested patients, N=93).Fig 3. Genetic Findings by Initial Presentation Type:
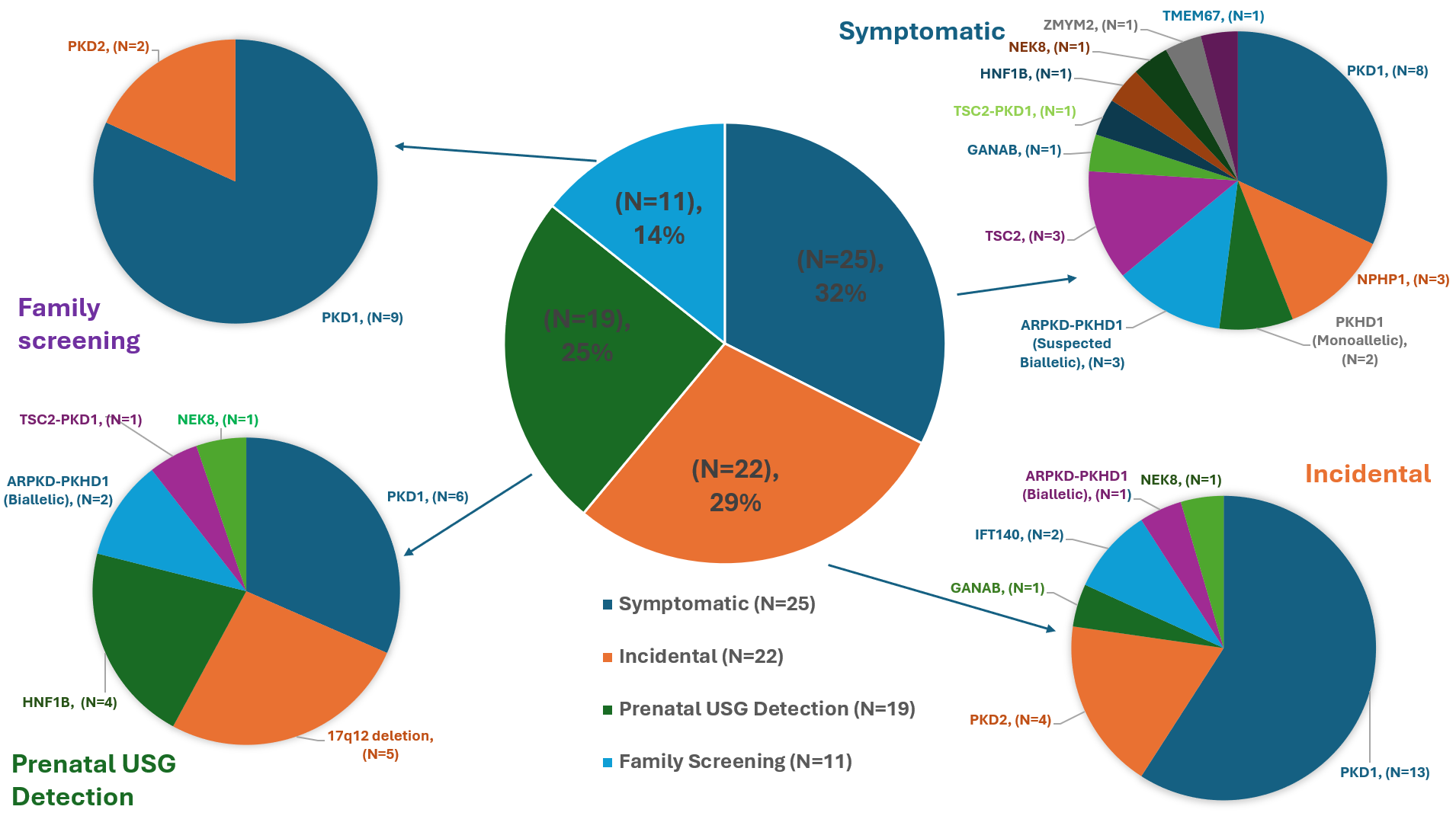 The central pie chart shows the distribution of resolved cases by initial presentation type: Symptomatic (32%), Incidental (29%), Prenatal Ultrasound Detection (25%), and Family Screening (14%). Each surrounding pie chart displays the distribution of causative genes for each presentation type. In Symptomatic cases, causative genes include PKD1, NPHP1, PKHD1 (monoallelic), suspected biallelic PKHD1 with ARPKD phenotype, TSC2, GANAB, TSC2-PKD1, HNF1B, NEK8, ZMYM2, and TMEM67. Incidental cases show PKD1, PKD2, GANAB, IFT140, PKHD1 (biallelic), and NEK8 as causative genes. Prenatal screening cases involve PKD1, 17q12 deletion, HNF1B, PKHD1 (biallelic), TSC2-PKD1, and NEK8. Family screening cases primarily feature PKD1 and PKD2.
The central pie chart shows the distribution of resolved cases by initial presentation type: Symptomatic (32%), Incidental (29%), Prenatal Ultrasound Detection (25%), and Family Screening (14%). Each surrounding pie chart displays the distribution of causative genes for each presentation type. In Symptomatic cases, causative genes include PKD1, NPHP1, PKHD1 (monoallelic), suspected biallelic PKHD1 with ARPKD phenotype, TSC2, GANAB, TSC2-PKD1, HNF1B, NEK8, ZMYM2, and TMEM67. Incidental cases show PKD1, PKD2, GANAB, IFT140, PKHD1 (biallelic), and NEK8 as causative genes. Prenatal screening cases involve PKD1, 17q12 deletion, HNF1B, PKHD1 (biallelic), TSC2-PKD1, and NEK8. Family screening cases primarily feature PKD1 and PKD2.Fig 1. (A) Disease-causing genes and their frequency in genetically resolved patients (N=77). (B) Disease-causing genes and their frequency in genetically resolved families (N=68).
.png) Variants were identified across a range of genes, including monoallelic or copy number changes in PKD1, PKD2, IFT140, NEK8, GANAB, PKHD1, TSC2, TSC2/PKD1, HNF1B, and ZMYM2, as well as biallelic variants in NPHP1 and TMEM67, and biallelic or suspected biallelic variants in PKHD1 associated with ARPKD phenotypes.
Variants were identified across a range of genes, including monoallelic or copy number changes in PKD1, PKD2, IFT140, NEK8, GANAB, PKHD1, TSC2, TSC2/PKD1, HNF1B, and ZMYM2, as well as biallelic variants in NPHP1 and TMEM67, and biallelic or suspected biallelic variants in PKHD1 associated with ARPKD phenotypes. Fig 2. Genetic Diagnostic Yield by Presentation Type:
 The y-axis categorizes initial presentation types: Family Screening, Prenatal Ultrasound (USG) Detection, Incidental, and Symptomatic. The x-axis shows the patient count. Solid-colored bars represent patients with resolved genetic diagnoses, with percentages indicating the resolution rate within each presentation type (Resolved patients, N=77). Dashed sections show the total number of patients tested per category (Tested patients, N=93).
The y-axis categorizes initial presentation types: Family Screening, Prenatal Ultrasound (USG) Detection, Incidental, and Symptomatic. The x-axis shows the patient count. Solid-colored bars represent patients with resolved genetic diagnoses, with percentages indicating the resolution rate within each presentation type (Resolved patients, N=77). Dashed sections show the total number of patients tested per category (Tested patients, N=93).Fig 3. Genetic Findings by Initial Presentation Type:
 The central pie chart shows the distribution of resolved cases by initial presentation type: Symptomatic (32%), Incidental (29%), Prenatal Ultrasound Detection (25%), and Family Screening (14%). Each surrounding pie chart displays the distribution of causative genes for each presentation type. In Symptomatic cases, causative genes include PKD1, NPHP1, PKHD1 (monoallelic), suspected biallelic PKHD1 with ARPKD phenotype, TSC2, GANAB, TSC2-PKD1, HNF1B, NEK8, ZMYM2, and TMEM67. Incidental cases show PKD1, PKD2, GANAB, IFT140, PKHD1 (biallelic), and NEK8 as causative genes. Prenatal screening cases involve PKD1, 17q12 deletion, HNF1B, PKHD1 (biallelic), TSC2-PKD1, and NEK8. Family screening cases primarily feature PKD1 and PKD2.
The central pie chart shows the distribution of resolved cases by initial presentation type: Symptomatic (32%), Incidental (29%), Prenatal Ultrasound Detection (25%), and Family Screening (14%). Each surrounding pie chart displays the distribution of causative genes for each presentation type. In Symptomatic cases, causative genes include PKD1, NPHP1, PKHD1 (monoallelic), suspected biallelic PKHD1 with ARPKD phenotype, TSC2, GANAB, TSC2-PKD1, HNF1B, NEK8, ZMYM2, and TMEM67. Incidental cases show PKD1, PKD2, GANAB, IFT140, PKHD1 (biallelic), and NEK8 as causative genes. Prenatal screening cases involve PKD1, 17q12 deletion, HNF1B, PKHD1 (biallelic), TSC2-PKD1, and NEK8. Family screening cases primarily feature PKD1 and PKD2.
