Nephrology 3
Session: Nephrology 3
598 - Identification of differentially expressed genes in Wilms tumor versus control samples
Sunday, April 27, 2025
8:30am - 10:45am HST
Publication Number: 598.5248
Michael T. Hsieh, University of Central Florida College of Medicine, Orlando, FL, United States; Brandon Molligoda, University of Central Florida College of Medicine, Altamonte Springs, FL, United States; Sarah Voskamp, University of Central Florida College of Medicine, Orlando, FL, United States; Jennifer S. Nelson, University of Central Florida College of Medicine, Orlando, FL, United States

Michael T. Hsieh (he/him/his)
Medical Student
University of Central Florida College of Medicine
Orlando, Florida, United States
Presenting Author(s)
Background: Wilms tumor (WT), or nephroblastoma, is a prevalent renal malignancy in children, with approximately 600 new cases diagnosed annually in the United States, representing 5% of pediatric cancers. However, it is highly treatable with five-year survival rates >90% for children with favorable histology and appropriate treatment.
Objective: This study aimed to elucidate novel molecular underpinnings of WT through a comprehensive meta-analysis of publicly available digital samples, comparing WT and healthy kidney. We primarily aimed to identify differentially expressed genes (DEGs) and canonical pathways involved in development of WT, highlighting novel molecular targets for detection and therapeutic intervention.
Design/Methods: Using the Search Tag Analyze Resource for Gene Expression Omnibus (STARGEO), we extracted 155 WT and 37 healthy control kidney samples and identified DEGs between both cohorts. Gene expression data from both cohorts were then analyzed for biological significance using ingenuity pathway analysis (IPA) restricted to genes with a statistically significant difference (p < 0.05) and an absolute experimental log ratio >0.1 between cohorts.
Results: Our analysis identified 2,926 DEGs between WT and healthy control samples. Significantly upregulated genes included PRAME (log ratio = 2.944), IGLC1 (2.570), ADGRG2 (1.536), and significantly downregulated genes included SLC12A1 (-3.309), DEFB1 (-3.308), and DDN (-3.251). These genes have varying roles including cell growth regulation and immune response. IPA also identified top canonical pathways: activation of cell cycle checkpoints (z-score = 9.110), mitotic prometaphase (7.318), and nuclear cytoskeleton signaling pathway (2.605). The top upstream regulators were beta-estradiol (0.217), dexamethasone (-6.944), TGFB1 (-0.645), ERBB2 (5.926), and TP53 (-5.504). Hepatic injury (3.801), hydronephrosis (3.643), and atrophy of the kidney (3.146) are some of the top upregulated toxicity functions.
Conclusion(s): Leveraging the STARGEO platform delineated robust genomic signatures associated with Wilms tumor pathology by interpreting extensive genomic data. The meta-analysis highlighted significant changes in gene expression in Wilms vs. control samples. These insights contribute to a deeper understanding of molecular mechanisms driving Wilms tumor and underscore the potential for developing targeted therapies to improve patient outcomes, even in patients with unfavorable histology. Future research should focus on validating these findings and exploring the therapeutic efficacy of targeting these pathways in clinical settings.
Gene Candidates
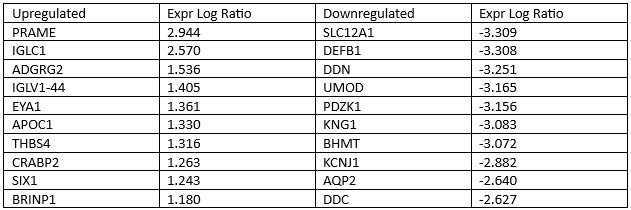 Table 1: Top 10 up and downregulated genes in Wilms tumor compared to healthy controls
Table 1: Top 10 up and downregulated genes in Wilms tumor compared to healthy controlsUpstream Regulator Dexamethasone
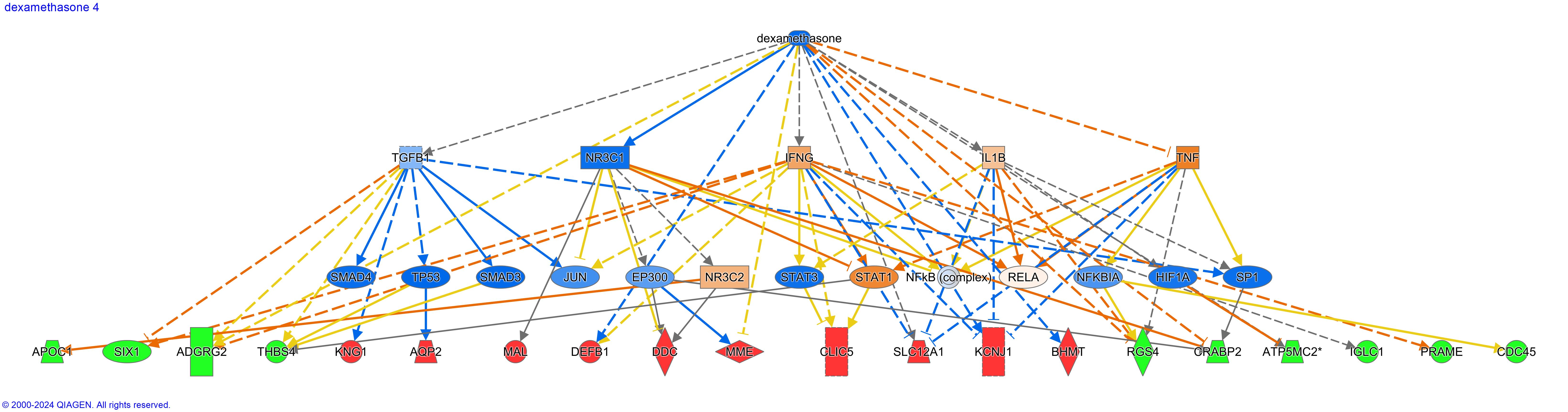 Figure 1. Dexamethasone, top upstream regulator predicted as inhibited with z-score -6.944, and downstream effects of inhibition of dexamethasone.
Figure 1. Dexamethasone, top upstream regulator predicted as inhibited with z-score -6.944, and downstream effects of inhibition of dexamethasone.Toxicity Function: Atrophy of Kidney
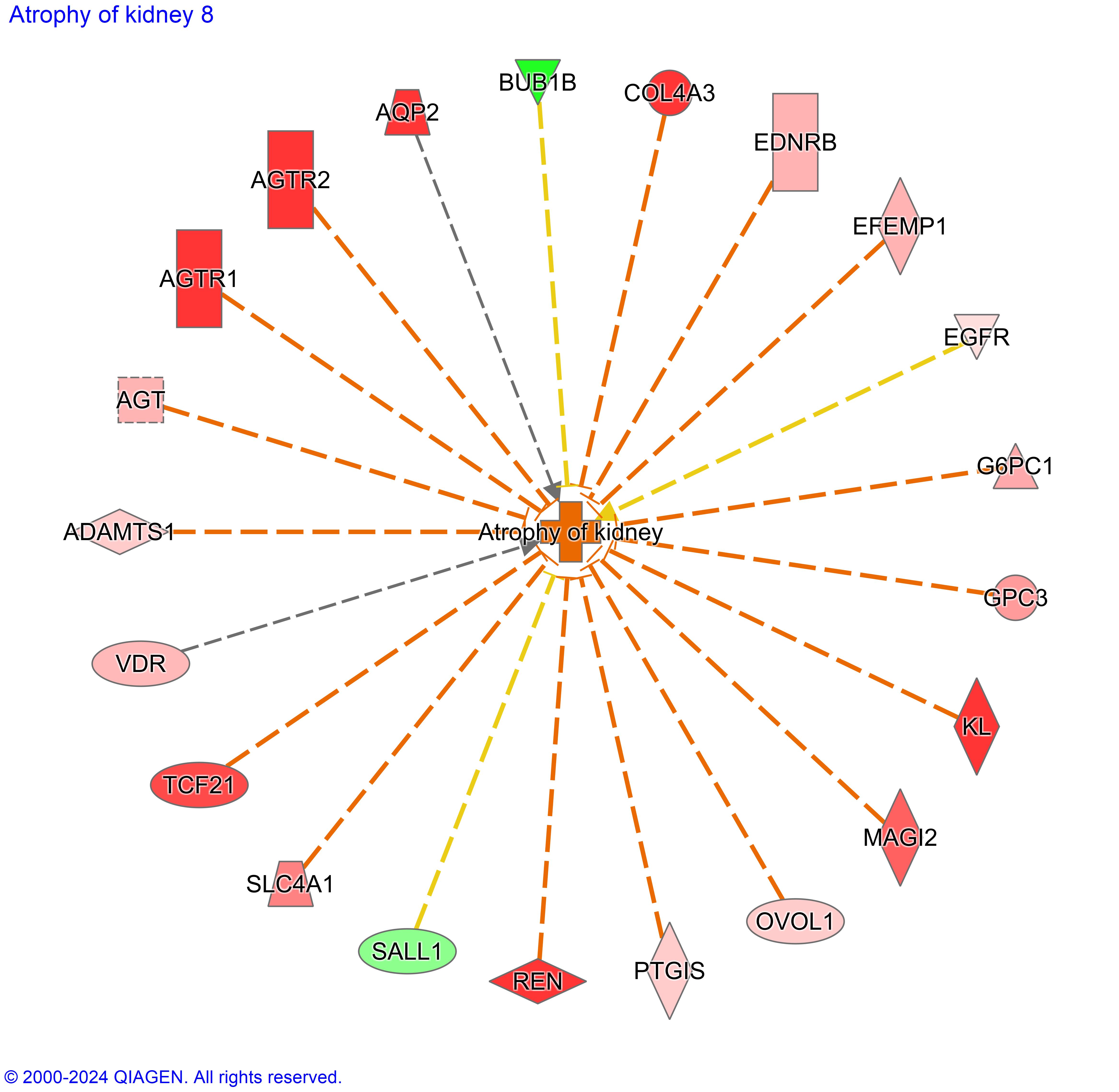 Figure 2. Atrophy of kidney, a top toxicity function associated with Wilms tumor. Atrophy of the kidney has predicted activation with a z-score of 3.146, suggesting the pattern of DEGs in WT contribute to renal atrophy.
Figure 2. Atrophy of kidney, a top toxicity function associated with Wilms tumor. Atrophy of the kidney has predicted activation with a z-score of 3.146, suggesting the pattern of DEGs in WT contribute to renal atrophy.
