Back
Background: Deep learning has significantly advanced brain MRI tissue segmentation in fetuses. Despite this, developing a model that performs optimally across a broad gestational age range (e.g., 17 to 41 weeks) remains challenging due to factors like rapid morphological changes and evolving tissue properties (e.g., water content, myelination).
Objective: To improve the performance of a previously validated fetal brain segmentation algorithm on younger fetuses (i.e., < 32 weeks gestational age) through hyperparameter search.
Design/Methods: In this new model, we increased the size of our training dataset from 65 to 124 fetal brain MRIs and widened the gestational age (GA) range from 23-39 to 19-39.4 weeks. We validated hyperparameters on 31 fetal datasets and tested on 3 fetuses with GAs of 23.1, 24.7, and 31.3 weeks. The initial hyperparameters were patch size = 48x48x48 (483), learning rate = 10-4, and epochs = 300. We then used different combinations of critical hyperparameters: patch size = 323 or 483, loss function: binary cross-entropy (BCE) or categorical cross-entropy (CCE), learning rate decay, early stopping mechanism, and data augmentation (DA). We evaluated the performance of models via visual inspection of outputs and comparing validation loss and Dice score (DS).
Results: We found that a smaller patch size reduced validation loss by a factor of ~10 (from 0.13037 to 0.01426). In contrast, learning rate decay and early stopping mechanism had little effect on reducing validation loss. We observed training and validation losses decayed over epochs, suggesting the successful training of the model (Fig. 1). We showed that CCE achieved better segmentations on the testing set (Fig. 2; DS=0.68±0.20 vs. 0.58±0.18 with BCE). DA also improved the segmentation accuracy (for BCE, DS=0.68±0.19 vs. 0.58±0.18; with vs. without DA). Combining CCE with DA resulted in failed segmentation in one fetus; this was successfully segmented using the original model (Fig. 2C). The new model successfully segmented the 23.2-week fetal brain; segmentation failed with the original model (Fig. 3).
Conclusion(s): Hyperparameter tuning on a larger dataset covering a broader GA range improved segmentation in two out of the three fetuses tested. The choice of loss function and data augmentation approach appeared to influence the quality of outputs the most; limited testing showed the potential of BCE + DA in fetuses < 24 weeks, but further studies that utilize increased sample sizes and perform extensive hyperparameter search are needed to build a more accurate U-Net-based segmentation algorithm generalizable across 2nd and 3rd trimester.
Figure 1. Training and validation curve
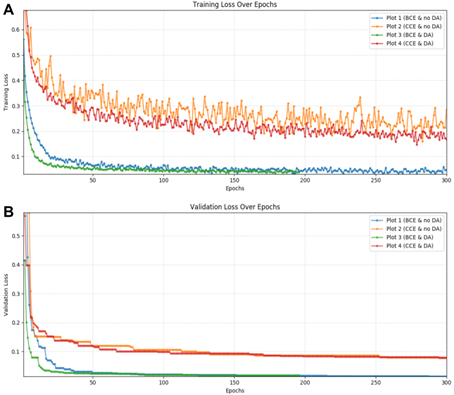 Plot 1: binary cross entropy (BCE) and no data augmentation (DA). Plot 2: categorical cross entropy (CCE) and no data augmentation. Plot 3: binary cross entropy and data augmentation. Plot 4: categorical cross entropy and data augmentation. (A) Training loss over epochs. (B) Validation loss over epochs.
Plot 1: binary cross entropy (BCE) and no data augmentation (DA). Plot 2: categorical cross entropy (CCE) and no data augmentation. Plot 3: binary cross entropy and data augmentation. Plot 4: categorical cross entropy and data augmentation. (A) Training loss over epochs. (B) Validation loss over epochs.
Figure 2. Segmentation quality in fetuses is influenced by loss function and data augmentation.
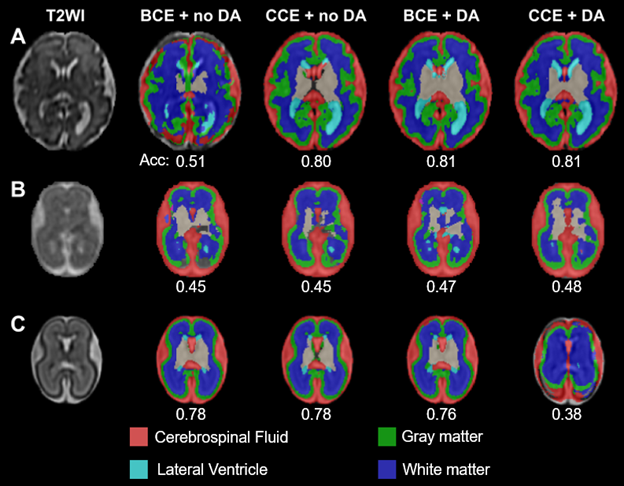 First column: T2-weighted image. Second column: binary cross entropy (BCE) and no data augmentation (DA). Third column: categorical cross entropy (CCE) and no data augmentation. Fourth column: binary cross entropy and data augmentation. Fifth column: categorical cross entropy and data augmentation. Acc: Segmentation accuracy compared to groundtruth segmentation results. GAs are 31.3, 23.2, and 24.7 weeks for A-C, respectively.
First column: T2-weighted image. Second column: binary cross entropy (BCE) and no data augmentation (DA). Third column: categorical cross entropy (CCE) and no data augmentation. Fourth column: binary cross entropy and data augmentation. Fifth column: categorical cross entropy and data augmentation. Acc: Segmentation accuracy compared to groundtruth segmentation results. GAs are 31.3, 23.2, and 24.7 weeks for A-C, respectively.
Figure 3. Improved segmentation of young fetuses.
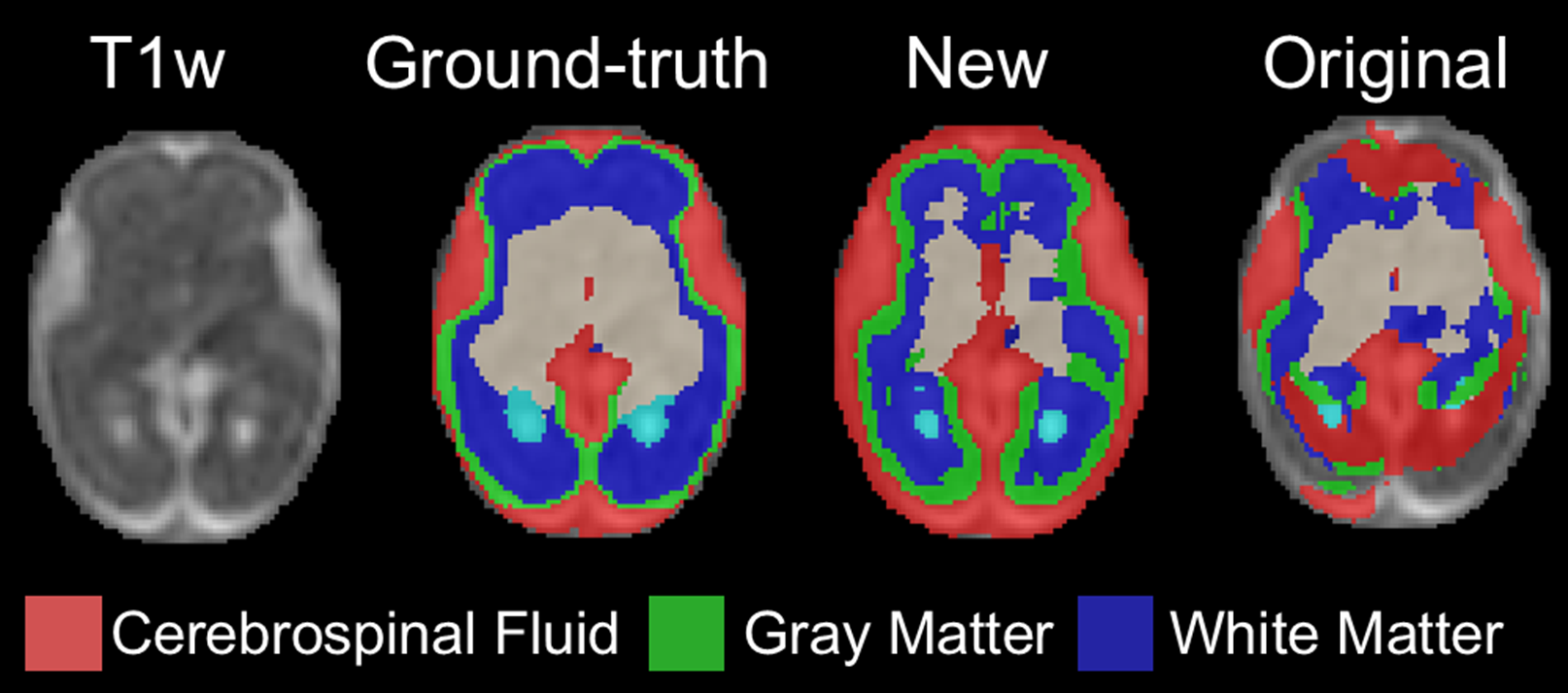 First column: T2-weighted image. Second column: Segmentation results with new model trained on young + older fetuses. Third column: Segmentation results with the original model trained on older fetuses.
First column: T2-weighted image. Second column: Segmentation results with new model trained on young + older fetuses. Third column: Segmentation results with the original model trained on older fetuses.
Neonatal Neurology 5: Fetal
Session: Neonatal Neurology 5: Fetal
671 - Optimizing hyperparameters of U-Net-based brain segmentation algorithms for late 2nd and early 3rd trimester fetuses
Sunday, April 27, 2025
8:30am – 10:45am HST
Jung-Hoon Kim, Children's National Health System, Washington, DC, United States; Arion Tripathi, Children's National Health System, Mclean, VA, United States; Catherine Limperopoulos, Children's National Health System, Washington DC, DC, United States; Nickie Andescavage, Children's National Health System, Washington, DC, United States; Josepheen De Asis-Cruz, Children's National Health System, Washington DC, DC, United States

Jung-Hoon Kim, PhD
Research Associate
Children's National Health System
Washington, District of Columbia, United States
Presenting Author(s)
Background: Deep learning has significantly advanced brain MRI tissue segmentation in fetuses. Despite this, developing a model that performs optimally across a broad gestational age range (e.g., 17 to 41 weeks) remains challenging due to factors like rapid morphological changes and evolving tissue properties (e.g., water content, myelination).
Objective: To improve the performance of a previously validated fetal brain segmentation algorithm on younger fetuses (i.e., < 32 weeks gestational age) through hyperparameter search.
Design/Methods: In this new model, we increased the size of our training dataset from 65 to 124 fetal brain MRIs and widened the gestational age (GA) range from 23-39 to 19-39.4 weeks. We validated hyperparameters on 31 fetal datasets and tested on 3 fetuses with GAs of 23.1, 24.7, and 31.3 weeks. The initial hyperparameters were patch size = 48x48x48 (483), learning rate = 10-4, and epochs = 300. We then used different combinations of critical hyperparameters: patch size = 323 or 483, loss function: binary cross-entropy (BCE) or categorical cross-entropy (CCE), learning rate decay, early stopping mechanism, and data augmentation (DA). We evaluated the performance of models via visual inspection of outputs and comparing validation loss and Dice score (DS).
Results: We found that a smaller patch size reduced validation loss by a factor of ~10 (from 0.13037 to 0.01426). In contrast, learning rate decay and early stopping mechanism had little effect on reducing validation loss. We observed training and validation losses decayed over epochs, suggesting the successful training of the model (Fig. 1). We showed that CCE achieved better segmentations on the testing set (Fig. 2; DS=0.68±0.20 vs. 0.58±0.18 with BCE). DA also improved the segmentation accuracy (for BCE, DS=0.68±0.19 vs. 0.58±0.18; with vs. without DA). Combining CCE with DA resulted in failed segmentation in one fetus; this was successfully segmented using the original model (Fig. 2C). The new model successfully segmented the 23.2-week fetal brain; segmentation failed with the original model (Fig. 3).
Conclusion(s): Hyperparameter tuning on a larger dataset covering a broader GA range improved segmentation in two out of the three fetuses tested. The choice of loss function and data augmentation approach appeared to influence the quality of outputs the most; limited testing showed the potential of BCE + DA in fetuses < 24 weeks, but further studies that utilize increased sample sizes and perform extensive hyperparameter search are needed to build a more accurate U-Net-based segmentation algorithm generalizable across 2nd and 3rd trimester.
Figure 1. Training and validation curve
 Plot 1: binary cross entropy (BCE) and no data augmentation (DA). Plot 2: categorical cross entropy (CCE) and no data augmentation. Plot 3: binary cross entropy and data augmentation. Plot 4: categorical cross entropy and data augmentation. (A) Training loss over epochs. (B) Validation loss over epochs.
Plot 1: binary cross entropy (BCE) and no data augmentation (DA). Plot 2: categorical cross entropy (CCE) and no data augmentation. Plot 3: binary cross entropy and data augmentation. Plot 4: categorical cross entropy and data augmentation. (A) Training loss over epochs. (B) Validation loss over epochs.Figure 2. Segmentation quality in fetuses is influenced by loss function and data augmentation.
 First column: T2-weighted image. Second column: binary cross entropy (BCE) and no data augmentation (DA). Third column: categorical cross entropy (CCE) and no data augmentation. Fourth column: binary cross entropy and data augmentation. Fifth column: categorical cross entropy and data augmentation. Acc: Segmentation accuracy compared to groundtruth segmentation results. GAs are 31.3, 23.2, and 24.7 weeks for A-C, respectively.
First column: T2-weighted image. Second column: binary cross entropy (BCE) and no data augmentation (DA). Third column: categorical cross entropy (CCE) and no data augmentation. Fourth column: binary cross entropy and data augmentation. Fifth column: categorical cross entropy and data augmentation. Acc: Segmentation accuracy compared to groundtruth segmentation results. GAs are 31.3, 23.2, and 24.7 weeks for A-C, respectively.Figure 3. Improved segmentation of young fetuses.
 First column: T2-weighted image. Second column: Segmentation results with new model trained on young + older fetuses. Third column: Segmentation results with the original model trained on older fetuses.
First column: T2-weighted image. Second column: Segmentation results with new model trained on young + older fetuses. Third column: Segmentation results with the original model trained on older fetuses.
