Technology 1: AI/Machine Learning
Session: Technology 1: AI/Machine Learning
179 - Predicting Serum Phosphate Levels in Very Preterm Infants Using Machine Learning
Saturday, April 26, 2025
2:30pm - 4:45pm HST
Publication Number: 179.6327
Asbjorn S. Westvik, Oslo University Hospital, Eiksmarka, Akershus, Norway; Sissel Jennifer Moltu, Oslo University Hospital, Oslo, Oslo, Norway; Oliver Tomic, Norwegian University of Life Sciences, Ås, Akershus, Norway; Charlotte Tscherning, oslo university hospital, Oslo, Oslo, Norway

Asbjorn S. Westvik, MD (he/him/his)
Pediatrician
Oslo University Hospital
Eiksmarka, Akershus, Norway
Presenting Author(s)
Background: Very preterm infants are at risk of hypophosphatemia ( < 1.4 mmol/L), a condition associated with increased risk of neurodisability (aOR 1.74) and mortality (aOR 2.07). Risk factors include being small for gestational age (SGA), high protein supply and low phosphate intake, whereas optimal nutrition may prevent it. Frequent blood sampling for monitoring serum phosphate (P) levels are painful, increase the risk of infections and lead to iatrogenic anemia. Hypophosphatemia is associated with hypercalcemia and hypokalemia, which can be measured by blood gas analysis (BGA), requiring little blood volume.
Objective: Our objective was to develop predictive models that can estimate P levels based on nutrient intake data, BGA and patient information (birth z-score, current z-score, gestational age (GA) and SGA-status) in the 1st week of life. An optimal model should be able to estimate actual values, but also predict P levels ahead of time (12 and 24 h).
Design/Methods: Observational study based on data from the ImNuT-trial (NCT03555019), an RCT in 120 preterm infants born before GA 29. In total, 1567 complete BGAs were available from week one. From these, three datasets were prepared. In dataset A, BGA data were matched to available P levels taken within 60 min. of the BGA (n=418). In datasets B (n=211) and C (n=228), BGA data were matched to P levels taken 10-14 h and 22-26 h after the BGA, respectively. Each new dataset was merged with nutrient intake data and patient information. Outliers were not removed.
A selection of machine learning (ML) regression algorithms was evaluated using the Python scikit-learn package, including elastic net (ENET), gradient boosting, partial least squares regression, random forests, support vector machines and XGboost. Model performance was measured by the root mean squared error (RMSE) expressed in mmol/L. Validation was performed on an anonymous, historical dataset of very preterm infants.
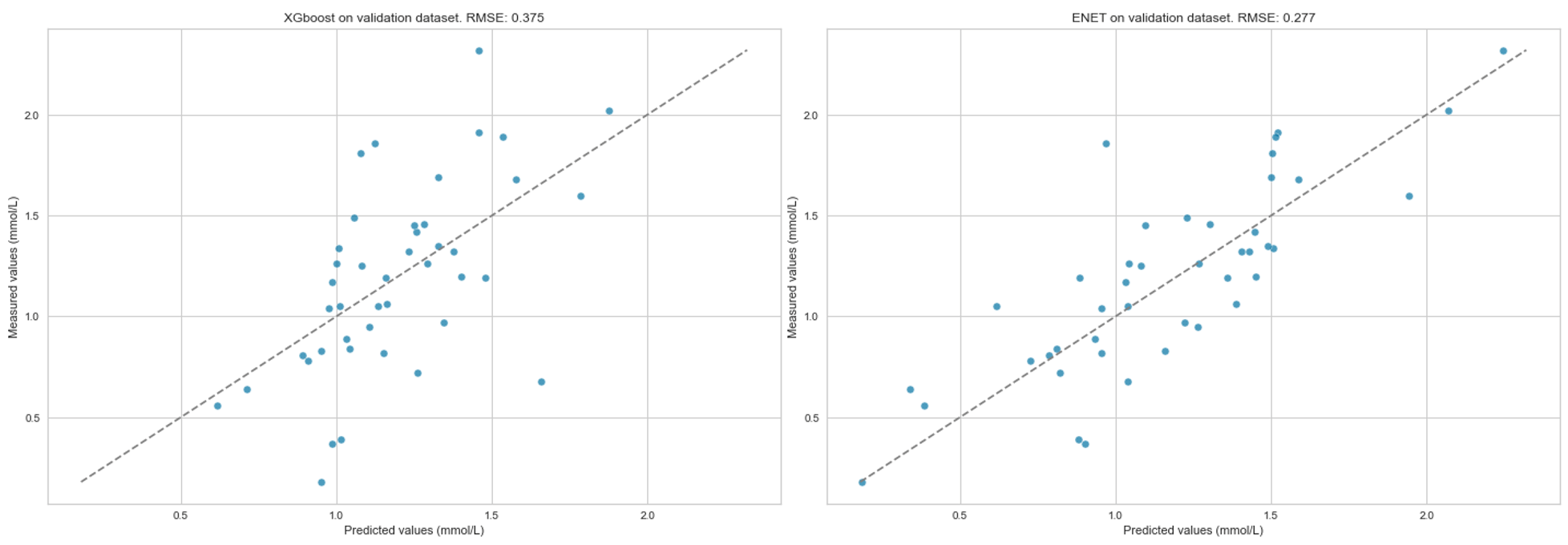
Results: Models built using ENET and XGBoost performed best. The models were able to predict the trend of P reasonably well in all three datasets (see Figures A-B). On A, RMSE was 0.28, on B 0.28 and on C 0.29. RMSE varied more on the validation dataset (0.28–0.38), with ENET performing the best (Figure C).
Conclusion(s): This study demonstrates that ML models, especially using ENET and XGBoost, can reasonably estimate actual and future P levels in very preterm infants. Though promising, these models require further refinement before they can be implemented clinically to minimize unnecessary interventions in low-risk infants and improve nutritional management in high-risk infants.
Figure A: Predicted versus measured serum phosphate level taken within 60 minutes of the blood gas analysis (Dataset A)
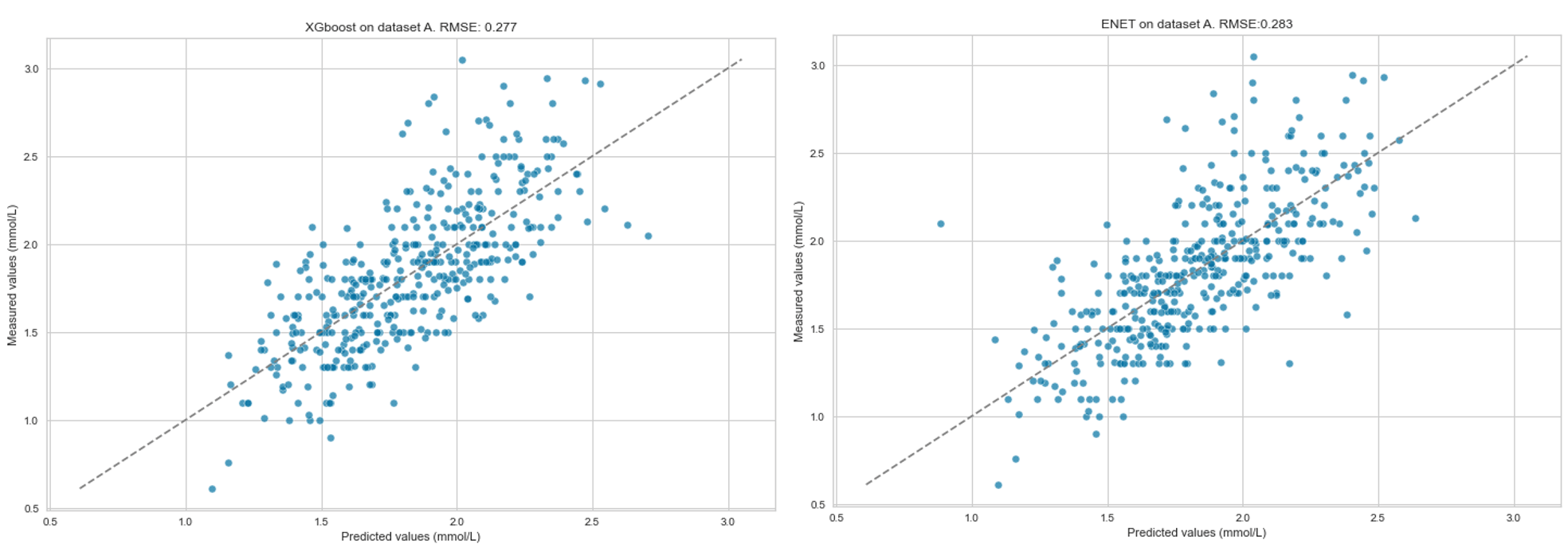
Figure B: Predicted versus measured serum phosphate level at 12 h (Dataset B) and 24 h (Dataset C) after the blood gas analysis
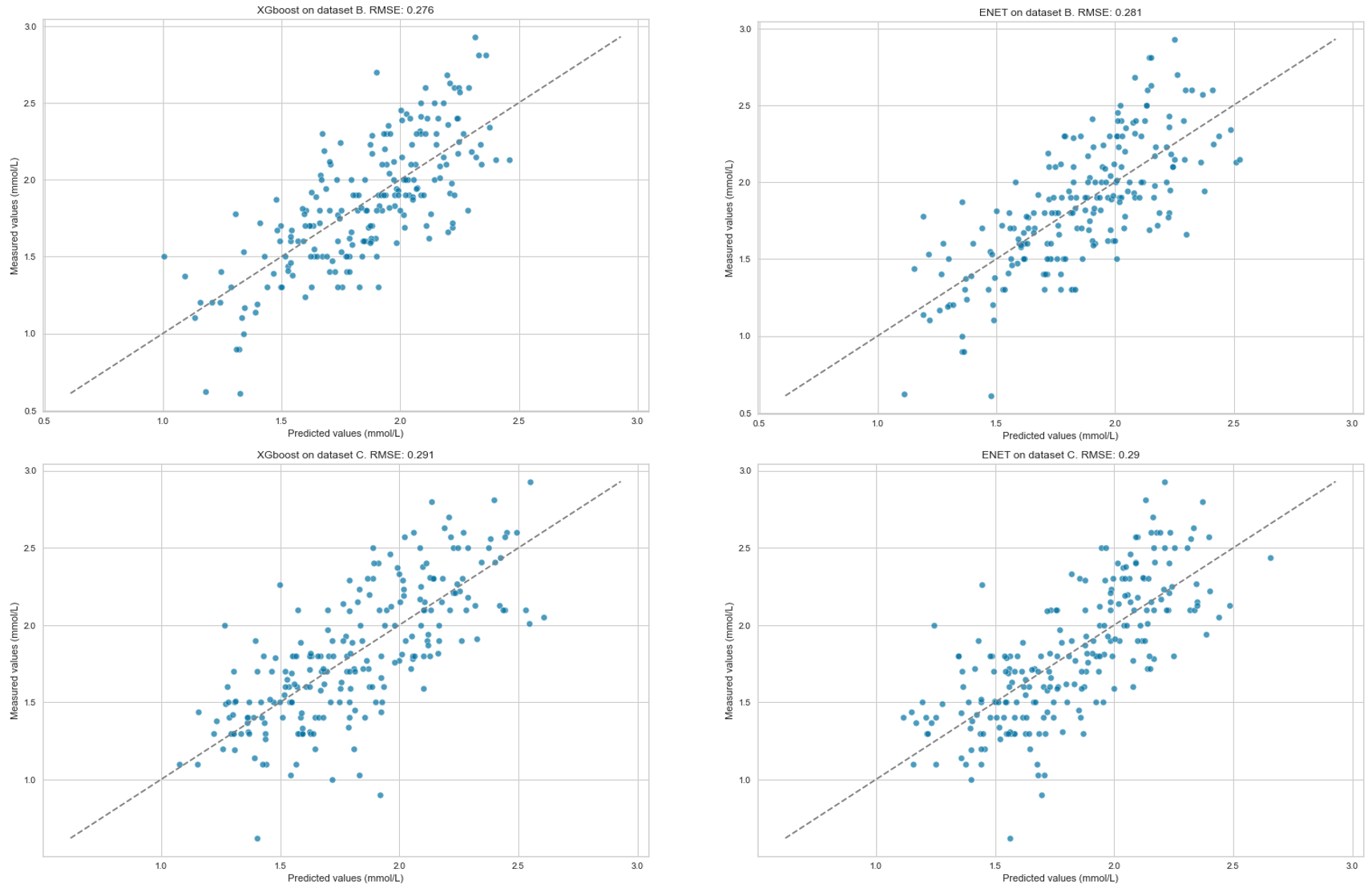
Figure C: Predicted versus measured serum phosphate level in the validation dataset.