Critical Care 3
Session: Critical Care 3
487 - Developing Automated Predictive Tool for Risk of Clinical Deterioration in Admitted Pediatric Patients Using Machine Learning
Monday, April 28, 2025
7:00am - 9:15am HST
Publication Number: 487.4463
Nga Tang, Loma Linda University Children's Hospital, SAN ANTONIO, TX, United States; Harsha Chandnani, Loma Linda University Children's Hospital, Loma Linda, CA, United States

Nga Tang, MD (she/her/hers)
Pediatric Intensivist
Loma Linda University Children's Hospital
SAN ANTONIO, Texas, United States
Presenting Author(s)
Background: Early recognition of pediatric patients’ clinical deterioration is crucial in improving patients’ outcomes. However, validated warning scores require manual input and additional effort in the context of the ever-growing complexity of electronic health records. We aimed to develop an automated predictive tool to detect clinical deterioration events using machine learning.
Objective: The primary objective of this study was to develop an automated tool to predict clinical deterioration (defined as rapid response or code event) in pediatric patients admitted to the hospital. We also aimed to identify EMR data points that impact the prediction model most significantly.
Design/Methods: This was a retrospective single center cohort study, using data from all pediatric patients between age > 37 weeks of gestation to 18 years of age admitted to the general wards, hematology-oncology unit, and step-down ICU of a tertiary pediatric center between July 2022 to March 2023. Data extracted was high dimensional mixed data, including demographics, vital signs, descriptive inputs of various assessments. Data reshaping was completed with pandas and ordinal encoder. Due to the heterogenous pathophysiology of pediatric patients, we opted for age-based grouping on vital signs. Synthetic Minority Oversampling Technique (SMOTE) used due to infrequent deterioration events in this population. Random Forest Classifier from scikit-learn was selected as the model of choice for supervised machine learning. Samples were divided 80-20% for model training and testing.
Results: Study population consisted of 3999 patients as 4952 unique encounters. Of these, 154 encounters had a rapid response or code event. We processed 37 features with mixed measurements, resulting in a sample size of 531,105. Oversampling increased the total sample size to 907,794. Our model’s performance was evaluated against a new sample size of 106,221. Our model showed great performance with AUROC score of 0.86, sensitivity of 0.71, true positive rate of 0.71, and false negative rate of 0.28. Features of age group, heart rate, gender, respiratory rate, SpO2, types of supplemental oxygen device, blood pressure, temperature had the highest impacts on prediction model.
Conclusion(s): Oversampling with random forest classifier is a practical method to develop an automated predictive tool for clinical deterioration, and it provides insights into important features at the bedside. This has potential to impact our understanding of clinical correlations in the era of precision medicine.
Confusion Matrix with Oversampling
.png) Our model's confusion matrix. Sensitivity of 0.71. True positive rate of 0.71. False negative rate of 0.28.
Our model's confusion matrix. Sensitivity of 0.71. True positive rate of 0.71. False negative rate of 0.28.ROC Curve with Oversampling
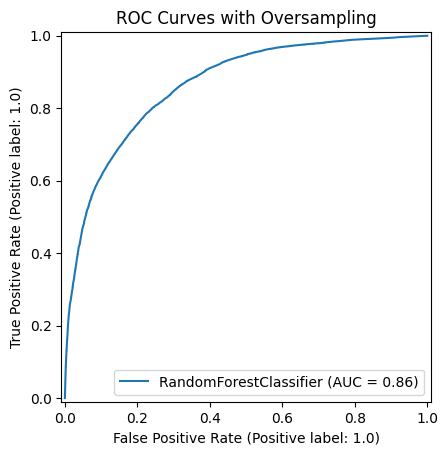 Our model's ROC Curve with oversampling, AUROC of 0.86.
Our model's ROC Curve with oversampling, AUROC of 0.86.Feature Importance Estimated by Random Forest Classifier
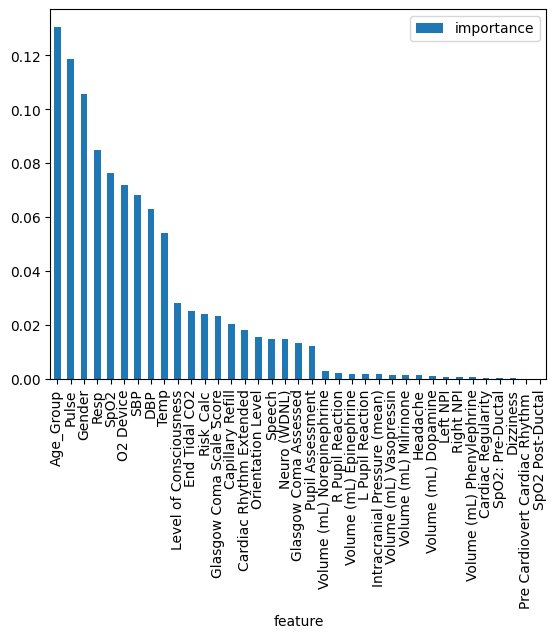 This shows which of the input features has the most impact on our model's predictions.
This shows which of the input features has the most impact on our model's predictions.
