Neonatal Pulmonology - Basic/Translational Science 2
Session: Neonatal Pulmonology - Basic/Translational Science 2
228 - Maternal Exposure to Diesel Exhaust Particles Exacerbates Neonatal Lung Injury Induced by Hyperoxia in Offspring Rats Through Increased Inflammation and Metabolic Disturbance
Friday, April 25, 2025
5:30pm - 7:45pm HST
Publication Number: 228.5751
Yushan Chang, Chi Mei Medical Center, Tainan, Tainan, Taiwan (Republic of China); Sheng-Yuan Ho, Tri-Service General Hospital, Taipei, Taipei, Taiwan (Republic of China); Hsiu-Chu Chou, Taipei medical university, Taipei, Taipei, Taiwan (Republic of China); Chung-Ming Chen, Taipei Medical University, Taipei, Taipei, Taiwan (Republic of China)

Yu-Shan Chang, MD, PhD (she/her/hers)
Attending physician
Chi Mei Medical Center
Tainan, Tainan, Taiwan (Republic of China)
Presenting Author(s)
Background: Air pollution poses a significant global health threat. Prenatal exposure to air pollution is linked to an increased risk of lung disease in offspring, although supporting animal studies are limited.
Objective: We aimed to investigate the effects of maternal diesel exhaust particles (DEP) exposure during pregnancy on offspring lung development and to determine whether prenatal DEP exposure exacerbates lung injury induced by postnatal hyperoxia.
Design/Methods: Pregnant Sprague-Dawley rats received daily intranasal instillations of DEP (500 ug) from gestational days 16 to 21. After birth, pups were assigned to room air (RA) or hyperoxia (85% oxygen). Four groups were established: PBS + RA, PBS + O2, DEP + RA, and DEP + O2. Pups were sacrificed at postnatal day 14 for lung examination.
Results: Offspring from DEP-exposed dams had lower birth weights compared to controls. Hyperoxia resulted in disrupted alveolarization, characterized by larger mean linear intercept lengths in the PBS + O2 group , which were further amplified in DEP-exposed pups. DEP exposure also increased levels of NF-κB p65, 8-OHdG, and inflammatory cytokines compared to controls, with postnatal hyperoxia exacerbating these effects. Metabolomics analysis of P14 lung tissues revealed significant metabolic profile differences between DEP+O2 and PBS+RA groups, with galactose, starch and sucrose, and amino sugar and nucleotide sugar metabolism as key disrupted pathways.
Conclusion(s): Maternal DEP exposure disrupts lung development and increases oxidative stress and inflammation in offspring, with postnatal hyperoxia further intensifying these effects.
Figure 1. Study design and body weight changes of offspring rats
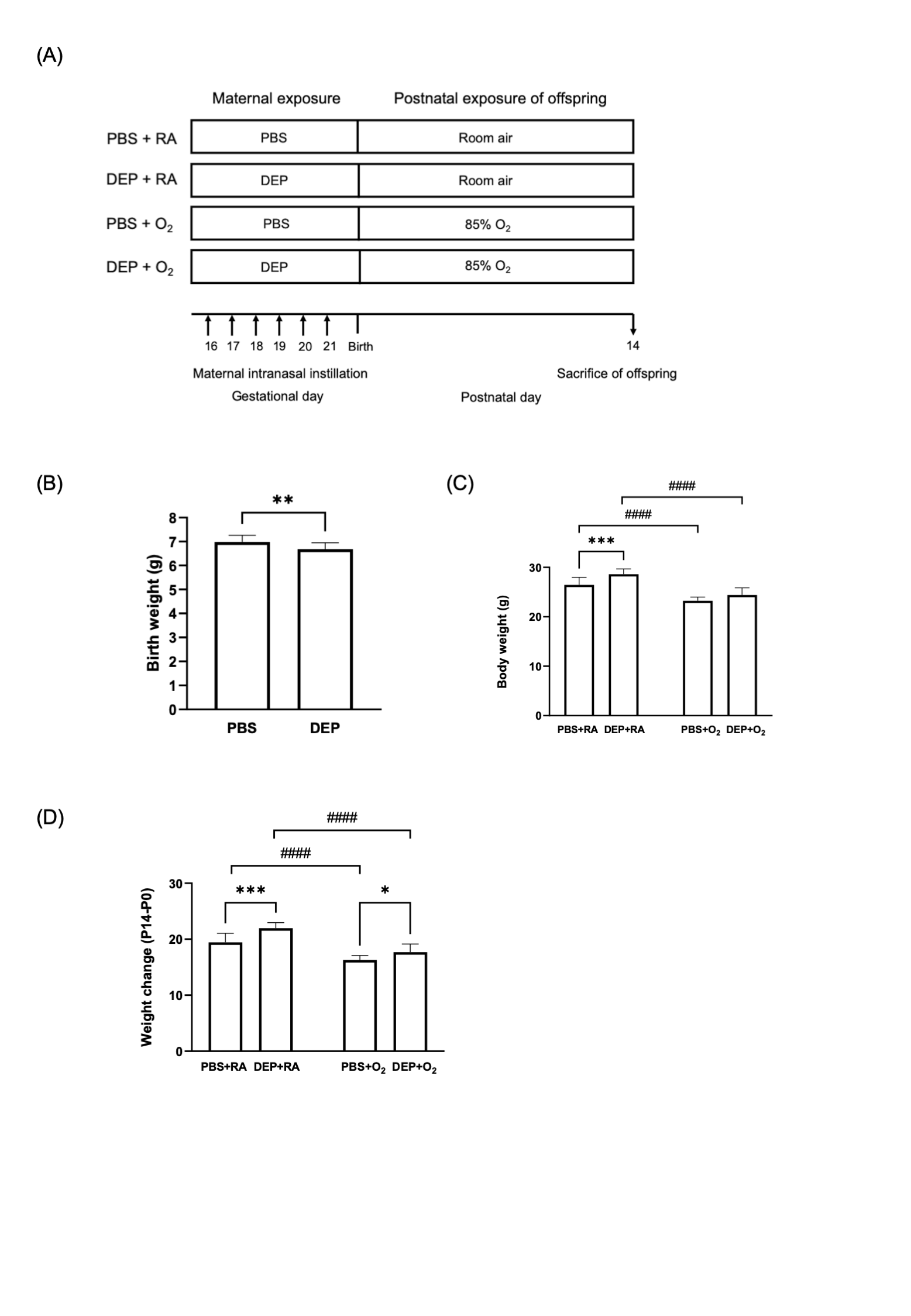 (A) Diagrammatic representation of the experimental design (B) Birthweight of offspring rats born to DEP-exposed and control (PBS) dams (C) Body weight of offspring rats at P14. (D) Weight changes (P14-P0) of offspring rats. The data are expressed as mean ± SD. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, #### p < 0.0001
(A) Diagrammatic representation of the experimental design (B) Birthweight of offspring rats born to DEP-exposed and control (PBS) dams (C) Body weight of offspring rats at P14. (D) Weight changes (P14-P0) of offspring rats. The data are expressed as mean ± SD. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, #### p < 0.0001Figure 2. The impact of maternal gestational DEP exposure and/or postnatal hyperoxia on offspring lung development.
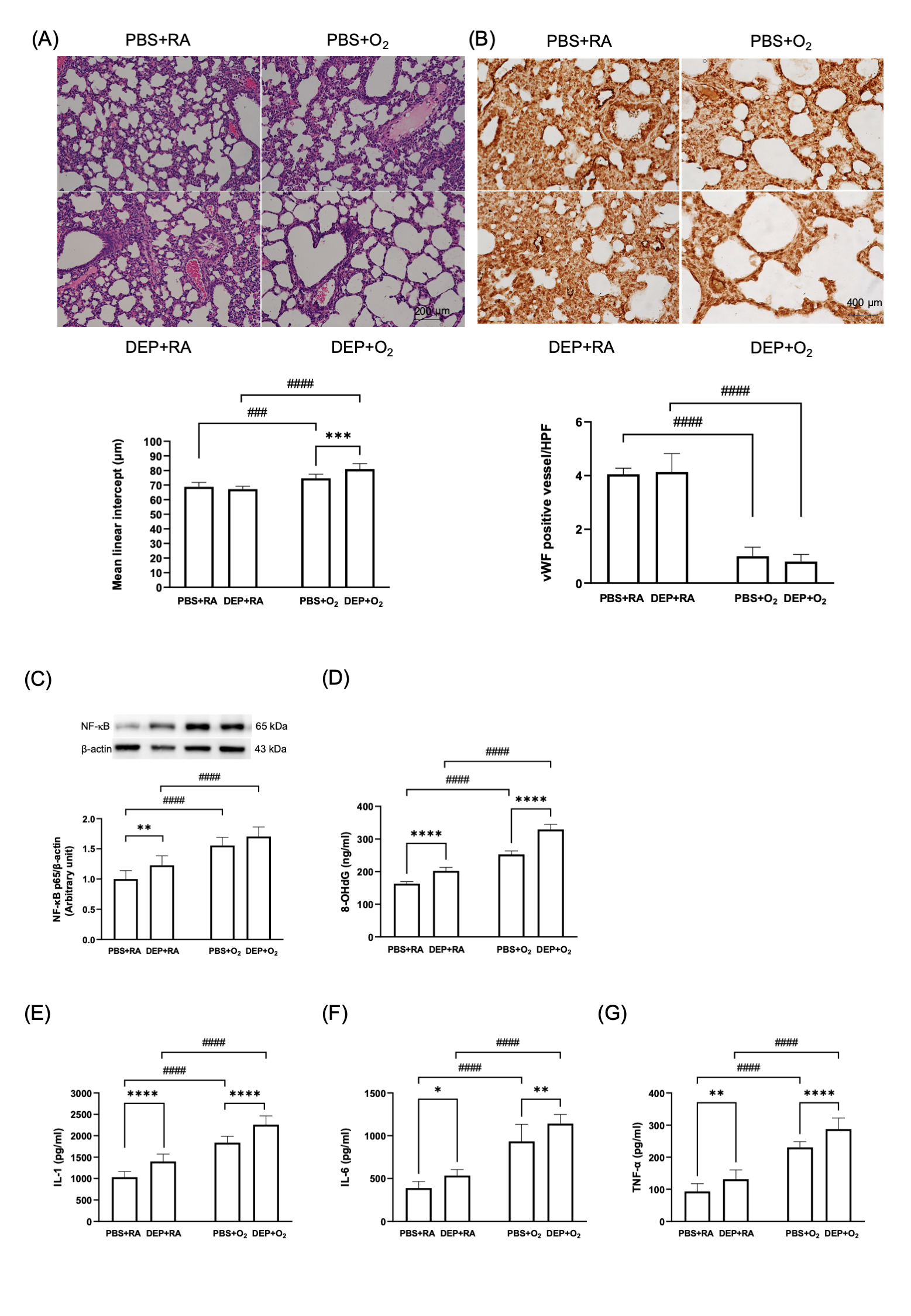 (A) Representative images of H&E stain of lung sections and quantification of mean linear intercept lengths. (B) vWF immunohistochemical stain (dark brown stain, indicated by arrows) of the lung and quantitative comparison of vWF expression. (C) Western blots of NF-kB protein and quantified results (D) 8-OHG level. Quantification of inflammatory cytokine levels including (E) IL-1, (F) IL-6 and (G) TNF-a. Data are expressed as mean ± SD. * p < 0.05, ** p < 0.01, *** p< 0.001, **** p < 0.0001, ### p < 0.001, #### p < 0.0001
(A) Representative images of H&E stain of lung sections and quantification of mean linear intercept lengths. (B) vWF immunohistochemical stain (dark brown stain, indicated by arrows) of the lung and quantitative comparison of vWF expression. (C) Western blots of NF-kB protein and quantified results (D) 8-OHG level. Quantification of inflammatory cytokine levels including (E) IL-1, (F) IL-6 and (G) TNF-a. Data are expressed as mean ± SD. * p < 0.05, ** p < 0.01, *** p< 0.001, **** p < 0.0001, ### p < 0.001, #### p < 0.0001Figure 3. Metabolic alterations in neonatal rat lungs on postnatal day 14.
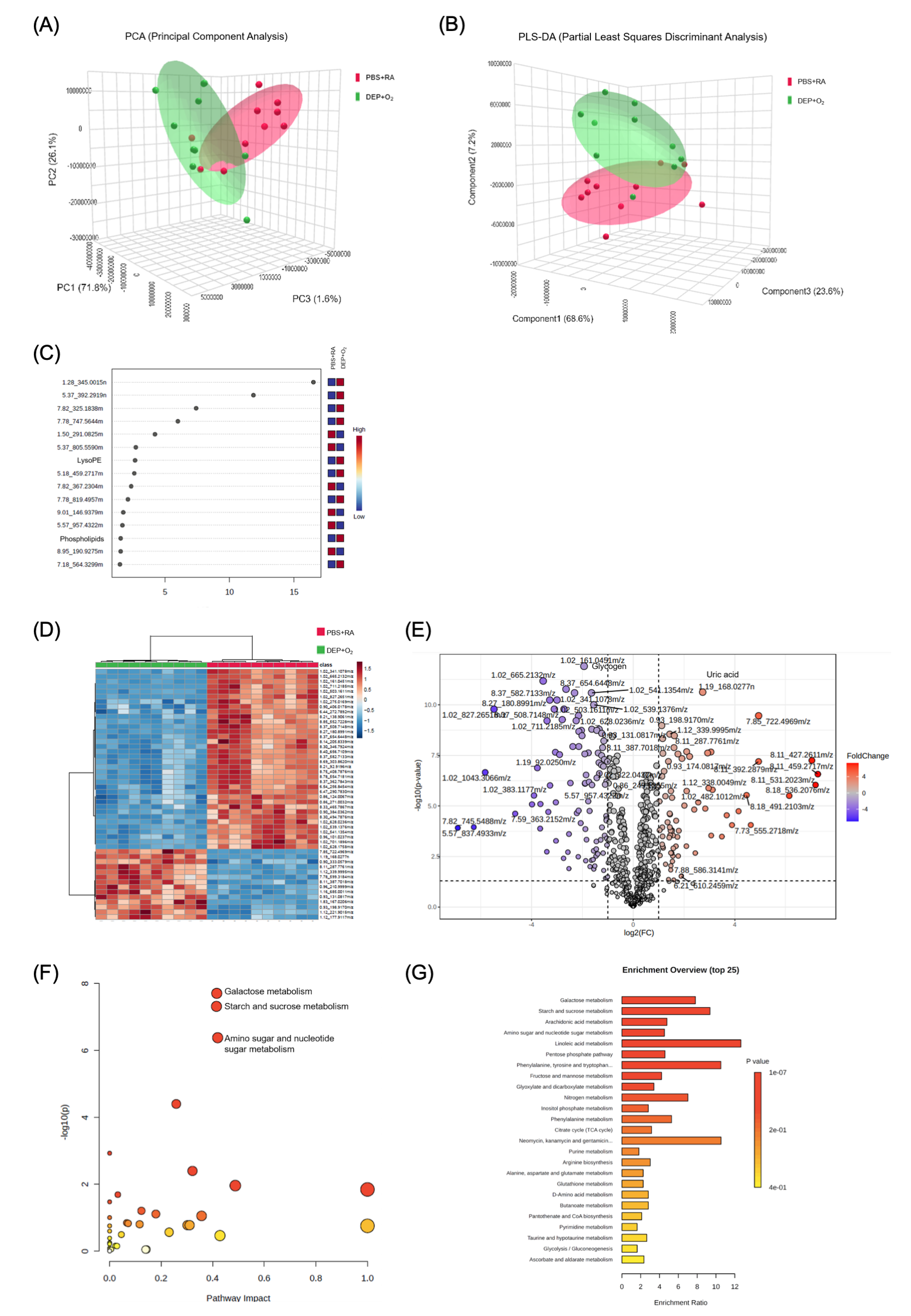 (A) Three-dimensional score plots from principal component analysis (PCA). (B) Partial least-squares discriminant analysis (PLS-DA) score plots for samples collected on postnatal day 14. Red circles represent the PBS+RA group, while green circles indicate the DEP+O2 group. (C) The compounds shown in the variable importance in projection scores are listed based on their ranking following false discovery rate adjustment for univariate analysis. The top 15 compounds that most significantly contributed to group differentiation were identified via PLS-DA and ordered by their variable importance in projection scores. The heatmap on the right illustrates the average intensities across the groups, where red indicates elevated metabolite levels and blue indicates reduced levels (n = 10). (D) Hierarchical clustering analysis. This analysis identified two distinct clusters that separate the PBS+RA and DEP+O2 groups in the lung. Clustering was conducted using Euclidean distance and Ward's linkage method. (E) Volcano plots illustrating the data from ultraperformance liquid chromatography–mass spectrometry (MS)/MS. The y-axis shows −log(p) values, while the x-axis represents log2 (fold change). Metabolites with significant alterations (fold change > 2, p < 0.05) are marked in blue and red, with gray dots indicating nonsignificant metabolites. (n = 10). Enrichment of metabolic pathways in the lungs of DEP+O2 rats displayed with (F) Bubble plot and (G) Bar chart, highlighting the metabolic pathways impacted in the DEP+O2 rat lungs on postnatal day 14 (n = 10).
(A) Three-dimensional score plots from principal component analysis (PCA). (B) Partial least-squares discriminant analysis (PLS-DA) score plots for samples collected on postnatal day 14. Red circles represent the PBS+RA group, while green circles indicate the DEP+O2 group. (C) The compounds shown in the variable importance in projection scores are listed based on their ranking following false discovery rate adjustment for univariate analysis. The top 15 compounds that most significantly contributed to group differentiation were identified via PLS-DA and ordered by their variable importance in projection scores. The heatmap on the right illustrates the average intensities across the groups, where red indicates elevated metabolite levels and blue indicates reduced levels (n = 10). (D) Hierarchical clustering analysis. This analysis identified two distinct clusters that separate the PBS+RA and DEP+O2 groups in the lung. Clustering was conducted using Euclidean distance and Ward's linkage method. (E) Volcano plots illustrating the data from ultraperformance liquid chromatography–mass spectrometry (MS)/MS. The y-axis shows −log(p) values, while the x-axis represents log2 (fold change). Metabolites with significant alterations (fold change > 2, p < 0.05) are marked in blue and red, with gray dots indicating nonsignificant metabolites. (n = 10). Enrichment of metabolic pathways in the lungs of DEP+O2 rats displayed with (F) Bubble plot and (G) Bar chart, highlighting the metabolic pathways impacted in the DEP+O2 rat lungs on postnatal day 14 (n = 10).
