Infectious Diseases 2: Bacterial infections
Session: Infectious Diseases 2: Bacterial infections
622 - Prevalence of Beta-Lactamase Antibiotic Resistance Genes in Neonatal E. coli Septicemia Isolates
Friday, April 25, 2025
5:30pm - 7:45pm HST
Publication Number: 622.6229
Eleana Cabello, University of Oklahoma Health Sciences, Oklahoma City, OK, United States; David W. Dyer, University of Oklahoma College of Medicine, Oklahoma City, OK, United States; Daniel P. Heruth, Children's Mercy, Kansas City, MO, United States; Joshua L. Wheatley, Children's Mercy Hospitals and Clinics, Olathe, KS, United States; Rangaraj A. Selvarangan, Childrens Mercy Kansas City, Kansas City, MO, United States; Susana Chavez-Bueno, Children's Mercy Kansas City and University of Missouri Kansas City School of Medicine, Kansas City, MO, United States

Susana Chavez-Bueno, MD (she/her/hers)
Professor of Pediatrics
Children’s Mercy Hospital
Kansas City, Missouri, United States
Presenting Author(s)
Background: E. coli is the dominant pathogen causing sepsis in the earliest neonatal period, with mortality up to 40% in preterm newborns. Widespread resistance to beta-lactam antibiotics has made treating these life-threatening infections increasingly challenging.
Objective: To characterize the prevalence of antimicrobial resistance (AMR) genes encoding beta-lactamases (BLA) among neonatal E. coli invasive isolates over time.
Design/Methods: Whole-genome (WGS) sequencing of 96 neonatal E. coli bacteremia isolates collected from 2003-2021 was performed with Illumina technology. Isolates were recovered from newborns with E. coli septicemia admitted to 2 tertiary hospitals in the US. After DNA extraction and sequencing, bacterial genomes were assembled de-novo using SKESA and annotated with Prokka. Contigs were screened using ABRIcate, and BLA-encoding genes identified using the AMRFinderPlus database (National Center for Biotechnology Information).
Results: Overall phenotypical nonsusceptibility rates to beta-lactams among all isolates were: Ampicillin (AMP) 60%, cefazolin 20%, ceftriaxone 6%, and piperacillin-tazobactam 6%. N=4 isolates produced extended-spectrum beta-lactamases (ESBLs) and were resistant to cefepime but were susceptible to piperacillin-tazobactam. We found no phenotypic carbapenem resistance.
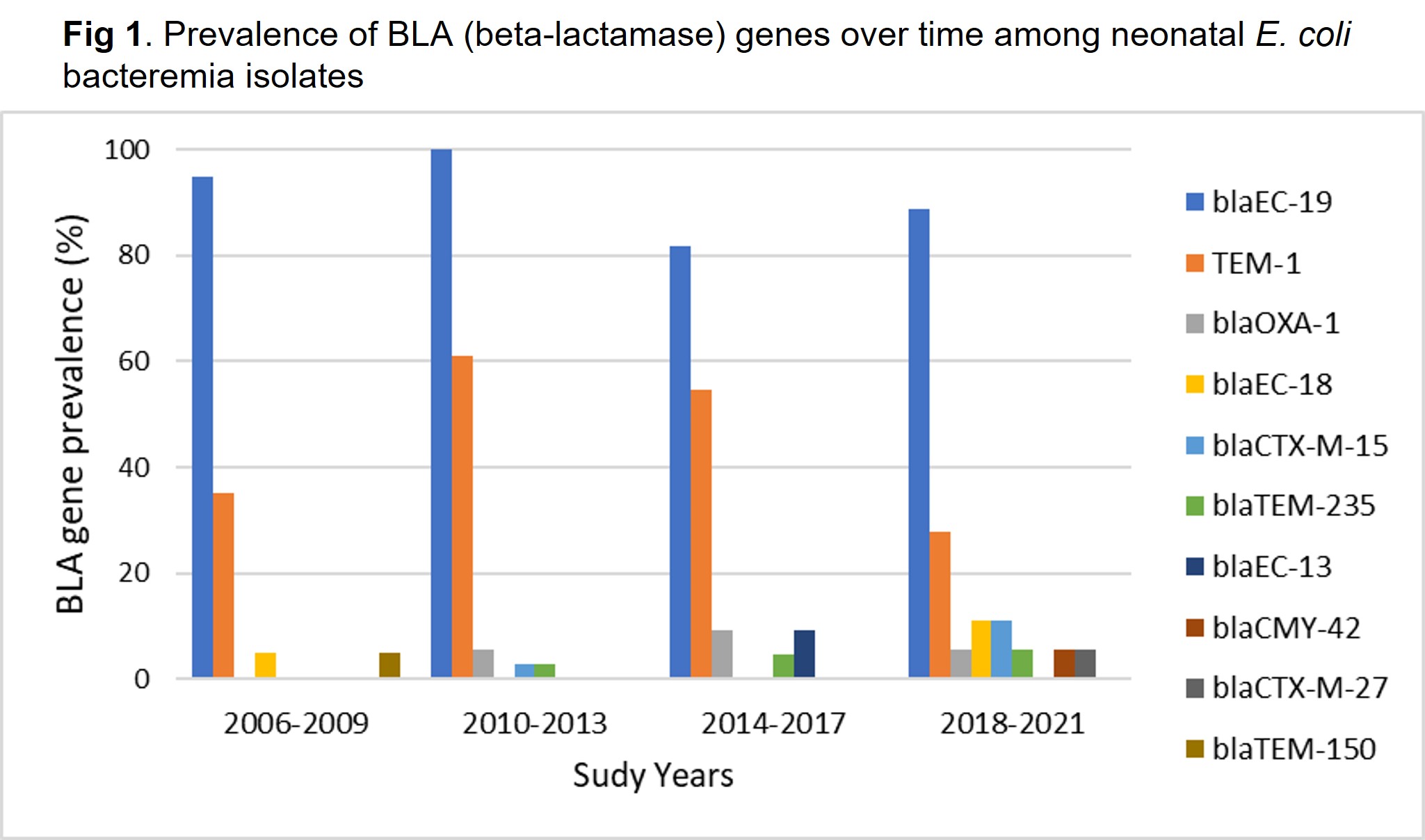
WGS analysis identified 10 beta-lactamase genes among all isolates. All isolates carried 1-3 BLA genes. N=40 (42%) carried 1, N=52 (54%) carried 2, and N=4 (4%) carried 3 BLA genes. The most prevalent were within Class C, and included blaEC-19, found in N=91 (95%) isolates (Table 1). Among the blaEC-19 carrying isolates, N=57 were AMP non-susceptible. N=46 (48%) of all isolates carried blaTEM-1, and N=45 were AMP non-susceptible. Of the ESBL-producing isolates, N=3 carried blaCTX-M-15 and N=1 carried blaCTX-M-27. blaOXA-1 was present in 5 isolates, including 2 of the ESBL isolates and 3 other isolates that were non-susceptible to AMP, but were susceptible to ceftazidime. Extended-spectrum BLA genes that emerged over time included blaCTX-M-15, blaCTX-M-27, and blaCMY-42 (Fig. 1).
Conclusion(s): We found a greater variety of BLA genes over time, which included broader spectrum resistance genes. Although some isolates did not show a resistance phenotype despite carrying BLA genes, most of them were resistant to first line antibiotics to treat neonatal E. coli sepsis. Close monitoring of genomic determinants of antibiotic resistance needs to continue, to design improved preventative and treatment strategies against these severe neonatal infections.
Table 1. Overall prevalence of BLA (beta-lactamase) genes among neonatal E. coli bacteremia isolates
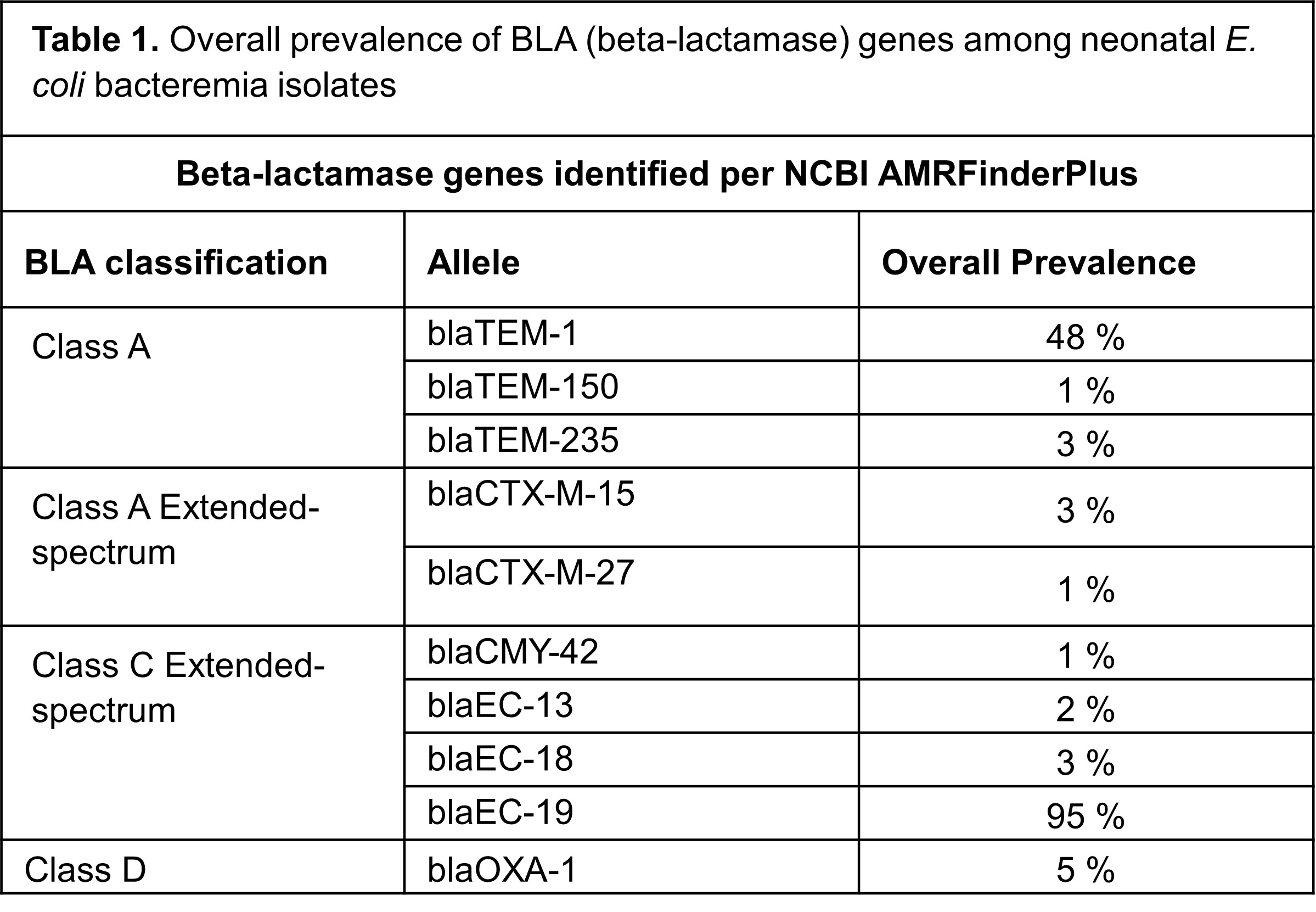
Fig 1. Prevalence of BLA (beta-lactamase) genes over time among neonatal E. coli bacteremia isolates