Neonatal Nephrology/AKI 1
Session: Neonatal Nephrology/AKI 1
004 - Downregulation of Renal Mitochondrial Energy Metabolism Genes in Adult Survivors of Murine Neonatal Sepsis may potentially Promote Chronic Kidney Dysfunction
Saturday, April 26, 2025
2:30pm - 4:45pm HST
Publication Number: 4.4821
Esther M. Speer, Renaissance School of Medicine at Stony Brook University, Stony Brook, NY, United States; Alexandra Khorasanchi, Renaissance School of Medicine at Stony Brook University, Stony Brook, NY, United States; Joyce Lin, Renaissance School of Medicine at Stony Brook University, Stony Brook, NY, United States; Robert Woroniecki, Stony Brook Children's Hospital, Stony Brook, NY, United States
Esther M. Speer, MD (she/her/hers)
Associate Professor of Pediatrics
Stony Brook University
Stony Brook, New York, United States
Presenting Author(s)
Background: Newborn sepsis (NS) is a major cause of mortality and multiorgan dysfunction including acute kidney injury (AKI). A high prevalence of chronic kidney disease (CKD) among former extremely preterm newborns has recently been reported, many of whom had a history of prior NS and/or AKI. How NS may promote the subsequent development of CKD remains however unclear. In order to elucidate potential mechanisms involved in the transition to CKD, we developed a murine model of NS to study its acute and chronic effects on renal tissue gene expression.
Objective: To investigate gene expression changes associated with lipopolysaccharide (LPS)-induced sterile sepsis in renal tissue of newborn mice as well as their adult survivors (AS).
Design/Methods: C57BL6/J mice (n=4-6 per group) were injected intraperitoneally with 2 µg per g body weight of LPS (from E. coli 0111:B4) on postnatal days (PN)1 and 2, or sterile saline for controls (CTL). Kidneys were harvested on PN3 or at 8 weeks of age to capture the acute and chronic sepsis-induced gene expression changes. Total RNA from homogenized renal tissue was extracted and mRNA purified and sequenced, employing Illumina next-generation sequencing. Differential gene expression was defined as |log2(FoldChange) ≥1 and Benjamini-Hochberg adjusted p-values < 0.05.
Results: As shown in the Volcano plot, many genes were upregulated during the acute phase of NS (Fig.1A), whereas AS primarily down-regulated genes compared to CTL (Fig.1B). The 10 most differentially expressed terms of the Gene Ontology (GO) analysis in renal tissue from NS vs CTL showed upregulation of leukocyte activation, chemotaxis, migration, and mitochondrial respiratory chain complex (Fig.2A). By contrast, GO terms of cellular components encoding mitochondrial and ribosomal structures (Fig.2B) as well as electron transfer activity were decreased in AS vs CTL. Significantly upregulated Reactome and KEGG (Kyoto Encyclopedia of Genes and Genomes) terms among NS included mitochondrial energy production and Toll-like receptor pathways (Table 1A), whereas collagen formation and extracellular matrix organization were suppressed. AS showed significant downregulation of mitochondrial energy metabolism including fatty acid oxidation, PPAR (peroxisome proliferator-activated receptor) and AMPK (AMP-activated protein kinase) signaling pathways (Table 1B).
Conclusion(s): While NS increased renal tissue expression of innate immunity and mitochondrial energy producing genes at the expense of extracellular matrix, oxidative energy metabolism genes were suppressed in kidneys of AS, thus potentially promoting the development of CKD.
Figure 1: Volcano plot of renal tissue gene expression among septic newborns (A) and 8-week old survivors of neonatal sepsis (B) vs their age-matched controls.
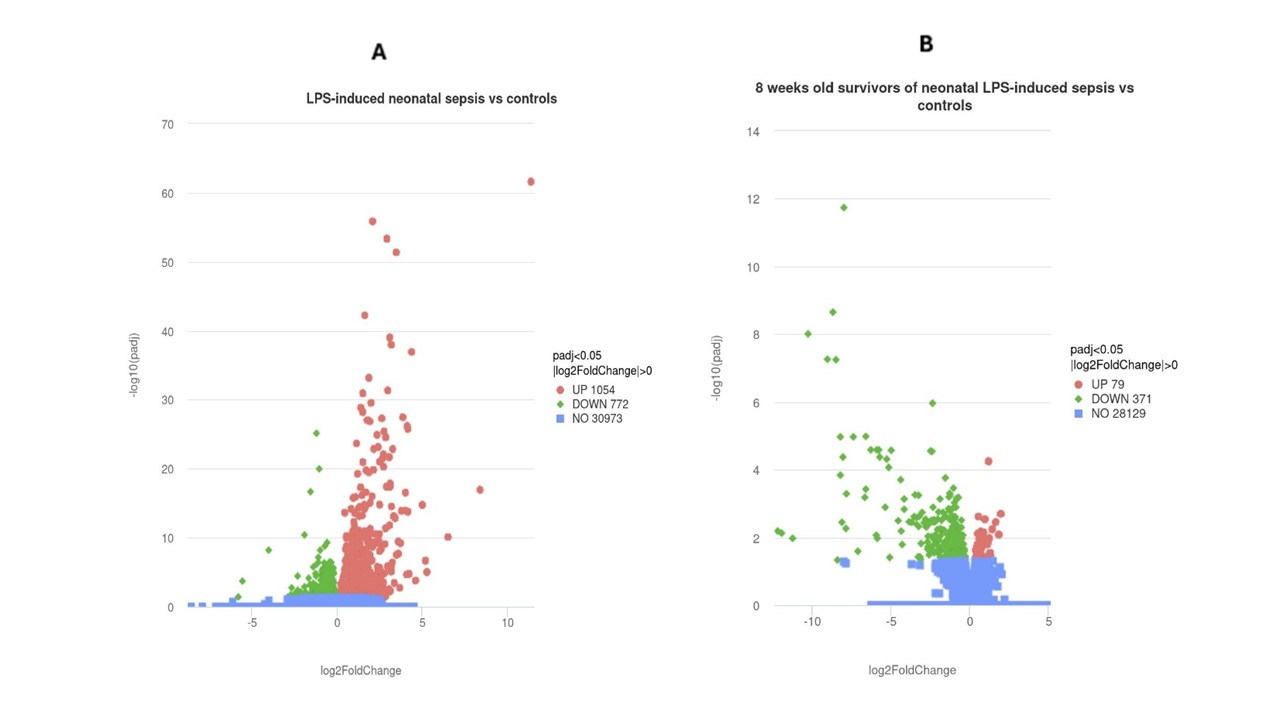 Upregulated genes (red), downregulated (green), unchanged gene expression (blue).
Upregulated genes (red), downregulated (green), unchanged gene expression (blue).Figure 2: Gene Ontology (GO) enrichment analysis of the 10 most differentially expressed genes among septic newborns (A) and 8-week old survivors of neonatal sepsis (B) vs their age-matched controls.
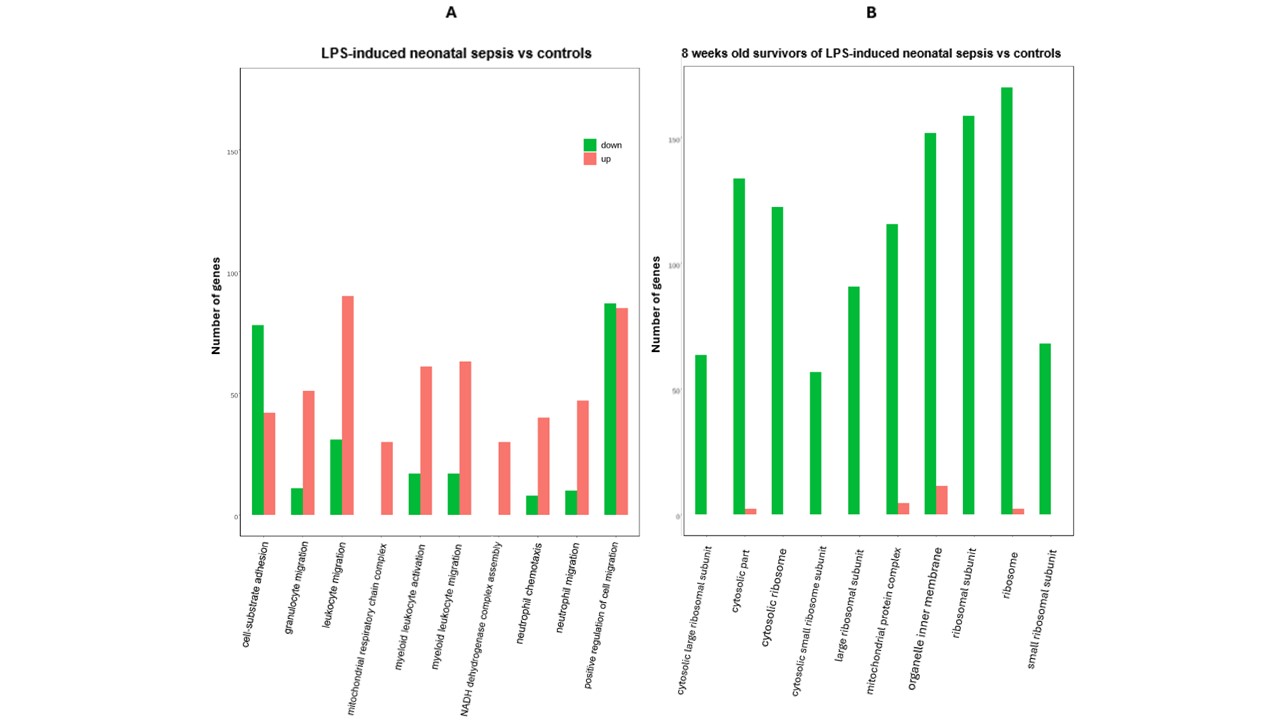 Upregulated genes (red), downregulated (green).
Upregulated genes (red), downregulated (green).Table 1: Differential gene expression KEGG and Reactome enrichment analysis of renal tissue from septic newborns (A) and 8-week old survivors of neonatal sepsis (B) vs controls.
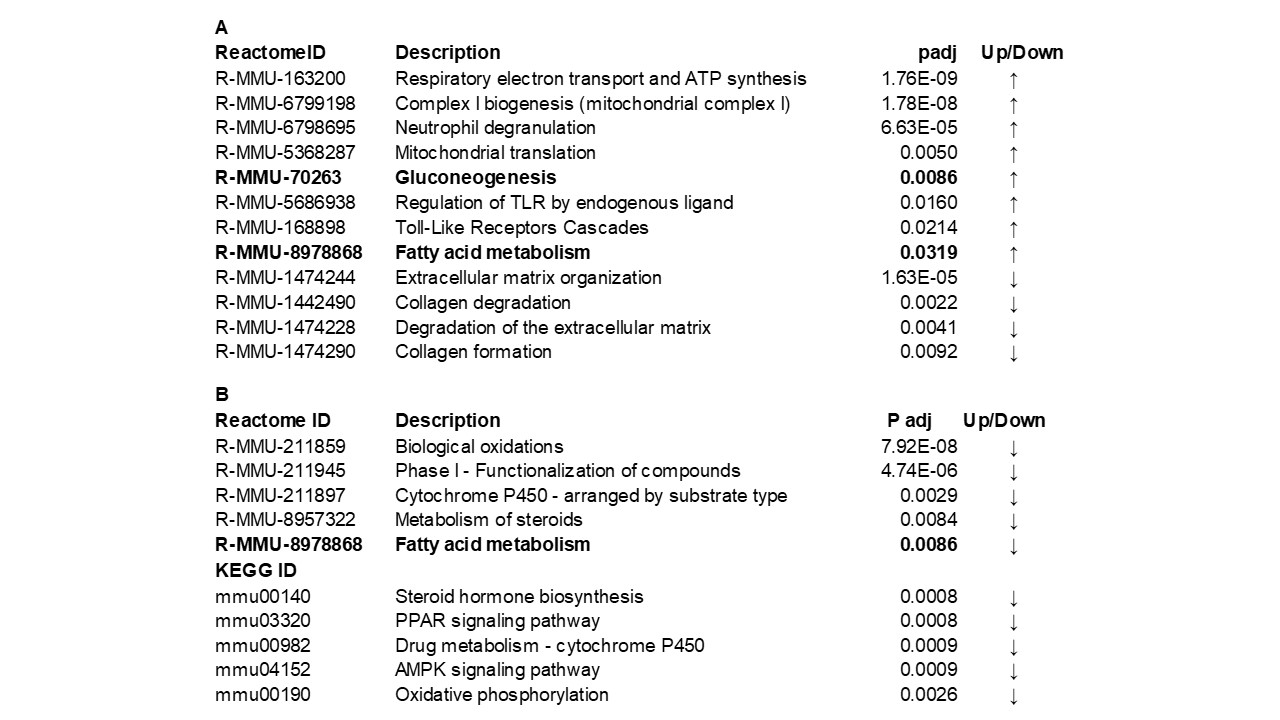 The arrows indicate up- or down-regulation of gene expression compared to controls; padj (Benjamini-Hochberg adjusted p-value).
The arrows indicate up- or down-regulation of gene expression compared to controls; padj (Benjamini-Hochberg adjusted p-value).
