Hematology/Oncology 1: Pediatric Oncology
Session: Hematology/Oncology 1: Pediatric Oncology
097 - Genetic differences between primary and metastatic Ewing Sarcoma: implications for targeted therapeutics
Saturday, April 26, 2025
2:30pm - 4:45pm HST
Publication Number: 97.4858
Carly Mitchell, University of Central Florida College of Medicine, Oviedo, FL, United States; Sarah Voskamp, University of Central Florida College of Medicine, Orlando, FL, United States; Hritik Nerusu, University of Central Florida College of Medicine, Orlando, FL, United States; Eddie Geagea, Nemours, Orlando, FL, United States; John Lovejoy, University of Central Florida College of Medicine, Belle Isle, FL, United States

Carly Mitchell, BS (she/her/hers)
MS3
University of Central Florida College of Medicine
Oviedo, Florida, United States
Presenting Author(s)
Background: Ewing sarcoma (ES) is the second most frequent bone tumor that can arise in the soft tissue or in the skeletal system, notably in the tibia, femur, pelvis, and ribs. Age ≥ 18 years, tumor size > 10 cm, radiotherapy alone, and no treatment were associated with increased risk of mortality. Ewing Sarcoma has been strongly linked to specific genetic influences, particularly a hallmark translocation between chromosomes 11 and 22, (t(11;22)(q24;q12)).
Objective: Herein we present an analysis of the genetic differences between primary and metastatic ES to characterize the genes associated with increased metastasis and identify therapeutic targets and molecular markers.
Design/Methods: Using the Search Tag Analyze Resource for NCBI’s Gene Expression Omnibus (STARGEO), 7 series were identified containing samples of Ewing Sarcoma metastasis (23 samples) and Ewing Sarcoma primary tumor (79 samples). Differentially expressed genes identified via STARGEO meta-analysis were restricted to p< 0.05 for statistical significance and absolute experimental log ratio greater than 0.1 for further analysis in Ingenuity Pathway Analysis (IPA), resulting in 753 analysis-ready molecules.
Results: Top upregulated genes in metastatic ES compared to primary ES tumors include ZNGE1, SFTA3, and SFTPC. Down regulated genes include OGN, CXCL14, and COMP which are genes that contribute to osteoblast differentiation, maintenance of macrophages, and proteins in the extracellular matrix. Extracellular matrix formation (z-score -3.000), assembly of collagen fibrils (-3.207), and integrin cell surface interactions (-2.324) are top canonical pathways with predicted inhibition. Upstream regulators include beta-estradiol, TGFB1, dexamethasone, TBX3, and ERBB2. The biological functions most activated in metastatic ES are transcription of DNA, transcription of RNA, and differentiation of epithelial tissue. However, the toxicity functions most activated include left ventricular hypertrophy and kidney damage.
Conclusion(s): Differentially expressed genes identified in metastatic ES enhance the understanding of potential for and mechanism of metastasis. The predilection of ES metastasis to the lung is supported by upregulation of genes encoding for surfactant proteins. Pro-SFTPB, the premature form of the upregulated gene SFTPB, may serve as a potential biomarker for metastatic ES as it is a known biomarker for osteosarcoma and lung cancer. The top causal network molecule of JBI-802 has significant antiproliferative effects and is currently in clinical trial phases 1 and 2 for tumor treatment and may be implicated as a therapeutic for metastatic ES.
Table 1. Top up and down regulated gene candidates in metastatic ES vs primary ES.
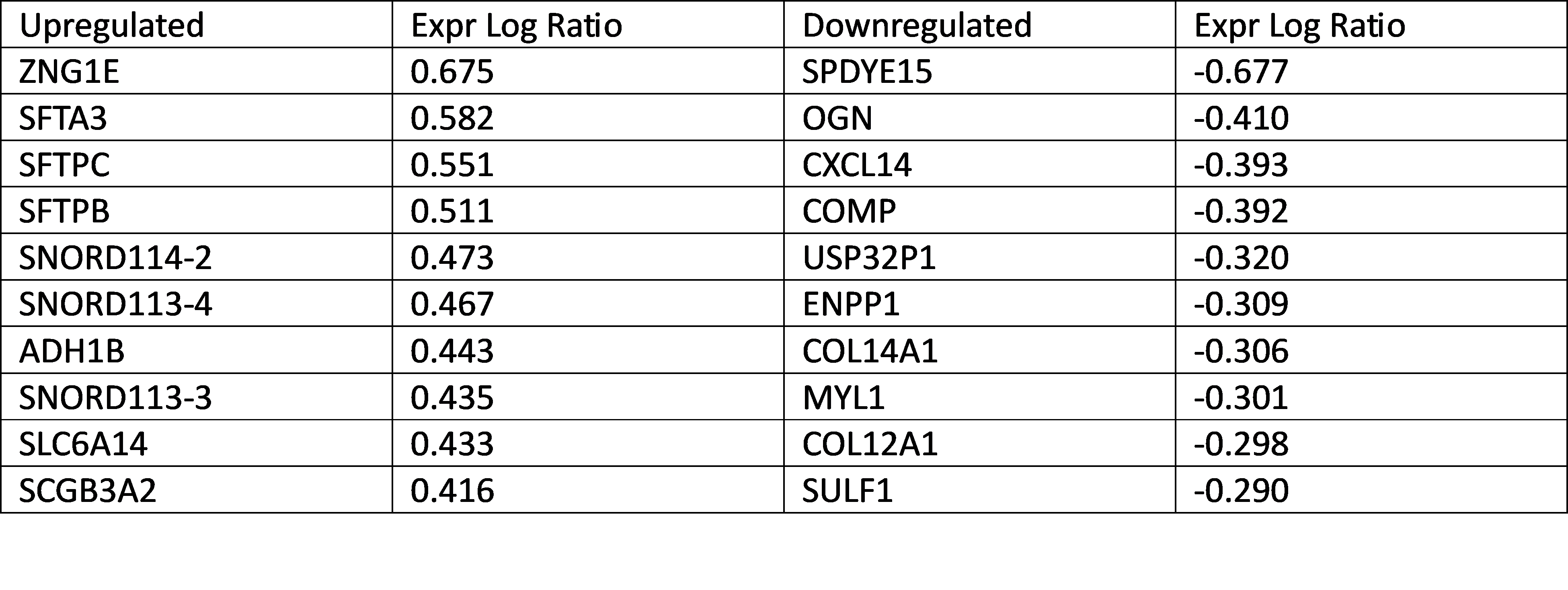
Figure 1. Beta-estradiol, top upstream regulator predicted as activated with corresponding downstream effects.
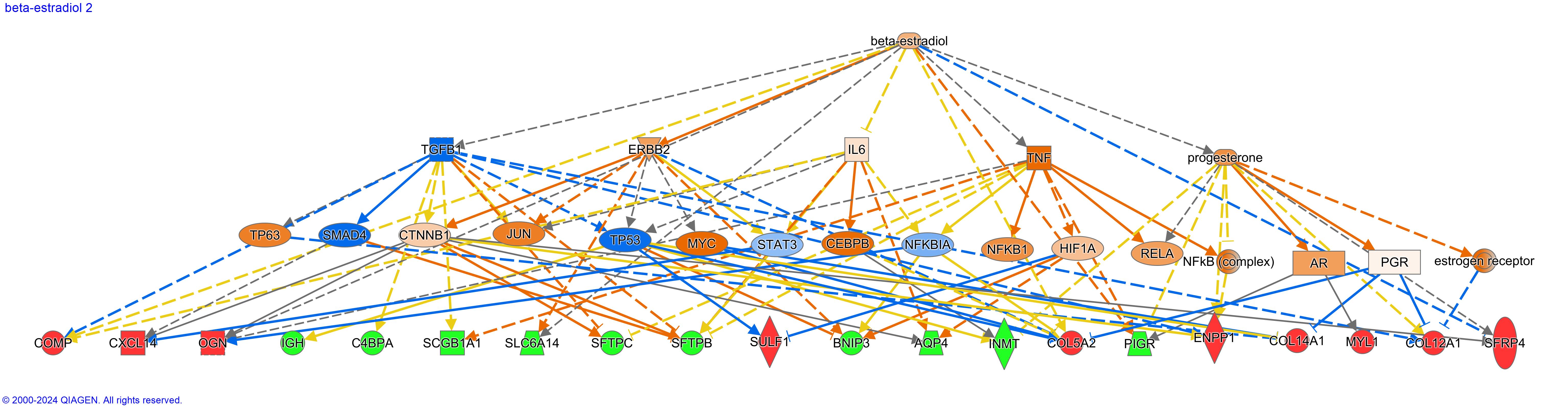 Upregulation of beta-estradiol implicates increased TNF and progesterone in metastasis of ES.
Upregulation of beta-estradiol implicates increased TNF and progesterone in metastasis of ES.Figure 2. JBI-802 as a top causal network.
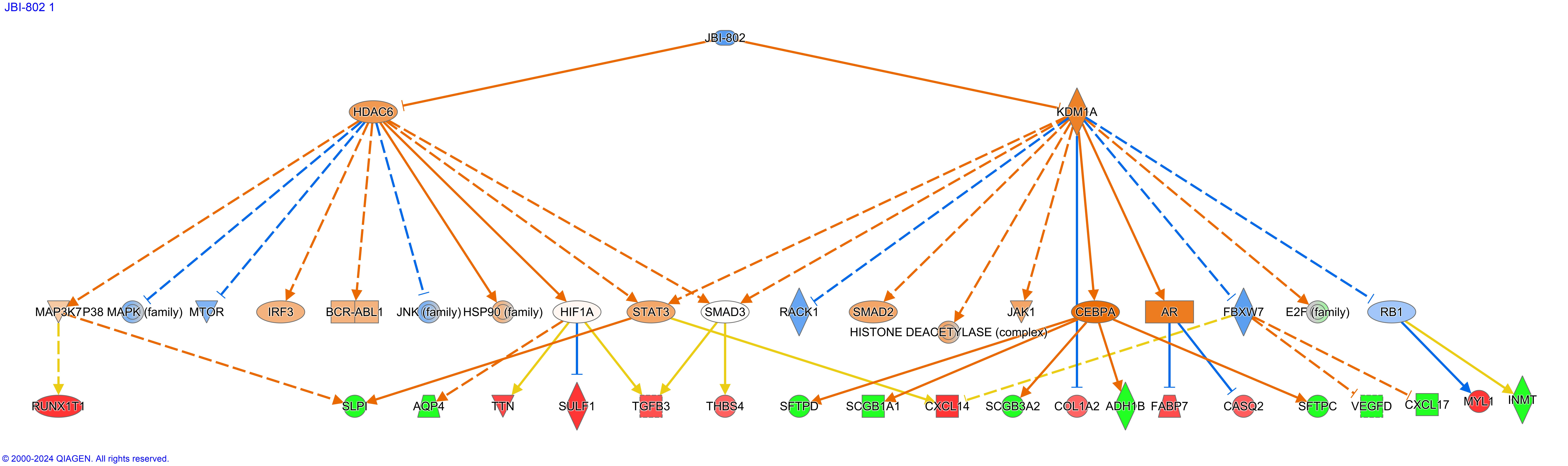 The JBI-802 network suggests potential for therapeutic drugs, warranting further investigation for metastatic ES. Downstream effects of JBI-802 are shown.
The JBI-802 network suggests potential for therapeutic drugs, warranting further investigation for metastatic ES. Downstream effects of JBI-802 are shown.
