Neonatal Fetal Nutrition & Metabolism 3
Session: Neonatal Fetal Nutrition & Metabolism 3
631 - Intrauterine Lipid Metabolism Modulates Placental Inflammatory Pathway in Mothers Affected by Obesity
Monday, April 28, 2025
7:00am - 9:15am HST
Publication Number: 631.5163
Ruggero Spadafora, Tufts Childrens Hospital, Weston, MA, United States; Jiayi Zhang, Tufts University, Boston, MA, United States; Perrie O'Tierney-Ginn, Tufts University School of Medicine, Boston, MA, United States

Ruggero Spadafora, MD (he/him/his)
Assistant Professor
Tufts Childrens Hospital
Weston, Massachusetts, United States
Presenting Author(s)
Background: The offspring of obese mothers have an increased risk of obesity, cardiovascular disease and non-alcoholic fatty liver disease (NAFLD). The placenta, which regulates both lipid metabolism and fetal-maternal immune homeostasis, is believed to play an important role in mediating and modulating these relationships. However, the molecular mechanism that in the placenta of mothers affected by obesity links altered lipid metabolism and dysregulated immunity remains unknown.
Objective: To correlate intrauterine metabolic traits with placental gene expression in mothers affected by obesity.
Design/Methods: Placentas (N=39) from mothers affected by obesity were obtained at birth. RNA-Seq was performed on villous tissue representing multiple placental biopsies. Whole genome correlation network analysis (WGCNA) was performed for placental coding genes to establish global correlations between placental genes that present similar patterns of expression (modules), and metabolic traits of the fetal-maternal dyad. Reactome, an open-source peer-reviewed database, was used to investigate the biological significance of the placental modules.
Results: Maternal Leptin, an important marker of the total body fat content, correlated positively with 2 modules (Red ρ=1; P-value =0.001; Brown ρ=0.98; P-value =0.02), while cord blood Free Fatty Acids (cbFFA) correlated positively with 1 module (Purple ρ=0.95; P-value < 0.05) (Fig.1). Strikingly, pathways analysis of the 3 modules showed a significant overrepresentation of every components of the Rho pathway such as RhoB and CDC42, DOCK8 and 11 and PAK2 and 4 that act, respectively, as activators and effectors of the Rho pathway. Rho is composed of signaling proteins that have crucial roles in triggering multiple immune functions with the involvement of effector proteins. Mutations of CDC42, a pathway represented 53 times in the modules is associated with immunodeficiency and immunological dysregulation
Conclusion(s): Here, we correlate placental gene expression with components of intrauterine lipid metabolism demonstrating significant positive modulation of the Rho inflammatory pathway in the placenta. Activation of Rho family components requires acceptance of an Acetyl-group from fatty acid donors (lipidation), suggesting a possible molecular link between fat dysregulation and inflammation in the placenta. Overall, these findings highlight the important role that lipid metabolism plays in modulating placental inflammation in maternal obesity. Further studies are required to investigate the placental inflammasome and the repercussions on fetal development.
Module-Metabolic Traits Correlation Heatmap
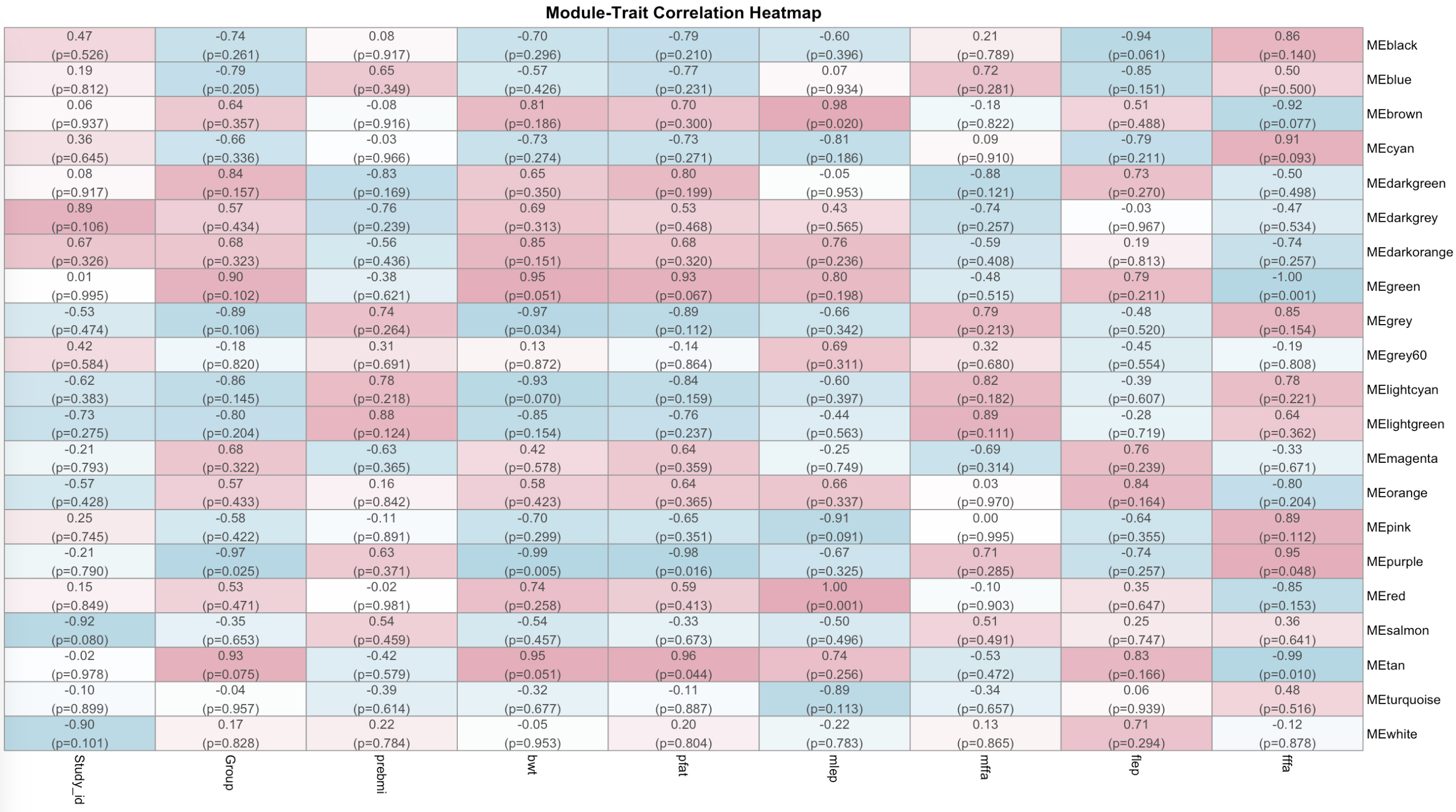 Using a Whole Genome Correlation Network Analysis (WGCNA) to study placental coding genes we established correlations between Maternal-Fetal Metabolic Traits and placental genes. Each correlation is characterized by a coefficient of correlation and a (P-value). Turquoise indicates negative correlation, Pink positive correlation. Notable negative correlations are: fffa with MEGreen and METan modules (-1, p=0.001; -0.99; p=0.01), notable positive correlations are: ffa with MEPurple and mlep with MERed and MEBrown modules (0.95, p=0.048; 1, p=0.001; 0.98, p=0.02).
Using a Whole Genome Correlation Network Analysis (WGCNA) to study placental coding genes we established correlations between Maternal-Fetal Metabolic Traits and placental genes. Each correlation is characterized by a coefficient of correlation and a (P-value). Turquoise indicates negative correlation, Pink positive correlation. Notable negative correlations are: fffa with MEGreen and METan modules (-1, p=0.001; -0.99; p=0.01), notable positive correlations are: ffa with MEPurple and mlep with MERed and MEBrown modules (0.95, p=0.048; 1, p=0.001; 0.98, p=0.02).fffa: fetal free fatty-acids; flep: fetal leptin, mffa: maternal fatty-acids, mlep: maternal leptin, pfat: neonatal adiposity, bwt: birth weight, preBMI: pre-pregnancy BMI
Module-Metabolic Traits Correlation Heatmap
 Using a Whole Genome Correlation Network Analysis (WGCNA) to study placental coding genes we established correlations between Maternal-Fetal Metabolic Traits and placental genes. Each correlation is characterized by a coefficient of correlation and a (P-value). Turquoise indicates negative correlation, Pink positive correlation. Notable negative correlations are: fffa with MEGreen and METan modules (-1, p=0.001; -0.99; p=0.01), notable positive correlations are: ffa with MEPurple and mlep with MERed and MEBrown modules (0.95, p=0.048; 1, p=0.001; 0.98, p=0.02).
Using a Whole Genome Correlation Network Analysis (WGCNA) to study placental coding genes we established correlations between Maternal-Fetal Metabolic Traits and placental genes. Each correlation is characterized by a coefficient of correlation and a (P-value). Turquoise indicates negative correlation, Pink positive correlation. Notable negative correlations are: fffa with MEGreen and METan modules (-1, p=0.001; -0.99; p=0.01), notable positive correlations are: ffa with MEPurple and mlep with MERed and MEBrown modules (0.95, p=0.048; 1, p=0.001; 0.98, p=0.02).fffa: fetal free fatty-acids; flep: fetal leptin, mffa: maternal fatty-acids, mlep: maternal leptin, pfat: neonatal adiposity, bwt: birth weight, preBMI: pre-pregnancy BMI

