Genomics/Epigenomics 1
Session: Genomics/Epigenomics 1
389 - Utilizing automated phenotype extraction for more timely whole genome sequencing
Friday, April 25, 2025
5:30pm - 7:45pm HST
Publication Number: 389.7044
Tanner Ellsworth, University of Utah, Salt Lake City, UT, United States; Emerson Lebleu, University of Utah School of Medicine, salt lake city, UT, United States; Phillip B. Warner, University of Utah School of Medicine, Salt Lake City, UT, United States; Kensaku Kawamoto, University of Utah, Salt Lake City, UT, United States; Peter Taber, University of Utah School of Medicine, Salt Lake City, UT, United States; Pinar Bayrak-Toydemir, University of Utah School of Medicine, Salt Lake City, UT, United States; Rong Mao, University of Utah School of Medicine, Salt Lake City, UT, United States; Paul Estabrooks, University of Utah School of Medicine, Salt Lake City, UT, United States; Martin Tristani-Firouzi, University of Utah School of Medicine, Salt Lake City, UT, United States; Sabrina Malone Jenkins, University of Utah, Salt Lake City, UT, United States

Tanner Ellsworth, MD, MPH (he/him/his)
Neonatal-Perinatal Medicine Fellow
University of Utah
Salt Lake City, Utah, United States
Presenting Author(s)
Background: Rapid whole genome sequencing (rWGS) is a valuable tool in diagnosis and treatment for rare genetic disorders in the NICU. Effective rWGS requires guidance using phenotype identification with Human Phenotype Ontology (HPO) terms. The standard process includes manual assignment of phenotypic terms and additional coding of HPO terms by the laboratory. Pheno+ is a novel application designed to reduce inefficiency by using interoperability standards and natural language processing to extract the phenotype and autogenerate HPO terms for clinical team curation.
Objective: This study compares differences in the time needed for phenotype identification and HPO term assignment when Pheno+ is included in the process for novice and experienced clinicians.
Design/Methods: One novice and one experienced clinician in genetic testing completed clinical documentation review using both the standard process and Pheno+ on the same 8 patients. The standard process involved review of clinical documentation to produce a list of phenotype terms and phenotype conversion to HPO terms by two laboratory molecular geneticists. The Pheno+ process involved clinicians selecting documentation from which a list of HPO terms were generated along with contextual document snippets, followed by manual selection of terms to keep, discard, or add. Four of the cases were reviewed using each approach two weeks apart. Time, notes reviewed, phenotype terms, and HPO terms were documented. Data are reported as median with interquartile range. Statistical analysis was performed using the Mann-Whitney U test.
Results: Using Pheno+ significantly reduced time spent from 24.5 minutes to 9.5 minutes (median, p< 0.001) (Table 1). This remained significant when separated by clinician experience (Table 2). Using Pheno+ also produced a significantly higher number of HPO terms, from 14.0 to 28.5 (med, p< 0.001) compared to standard review. When separated by clinician experience level, this difference was significant for the novice (p < 0.001) though only approached significance for the experienced clinician (p=0.06). Though we expected a significantly higher number of clinical documents to be reviewed using Pheno+, we ultimately did not see a significant difference between the two methods (p=0.35).
Conclusion(s): Pheno+ can significantly improve the speed and number of HPO terms selection for more accurate sequencing analysis regardless of the genetic experience of the clinician. This addresses a critical need with the expansion of genetic testing in the NICU and the efficiency can lead to expedited diagnosis and treatment for neonates with rare genetic disorders.
Table 1
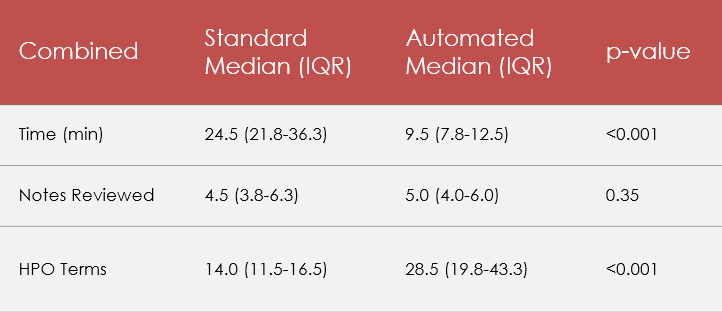 Standard vs. Pheno+ comparison, combined experience level
Standard vs. Pheno+ comparison, combined experience levelTable 2
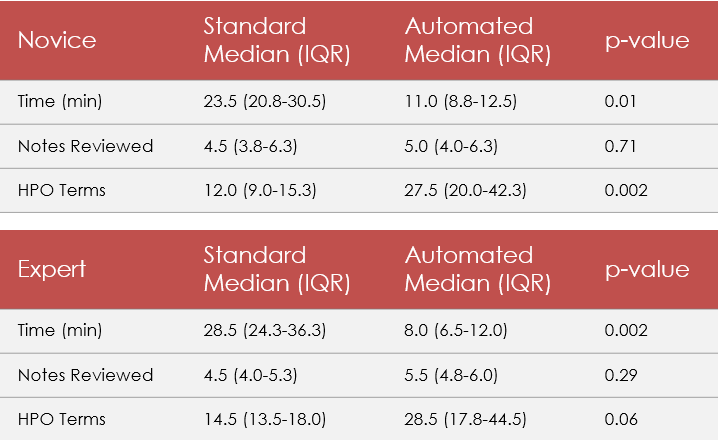 Standard vs. Pheno+ comparison, separated by clinician experience level
Standard vs. Pheno+ comparison, separated by clinician experience level
