Genomics/Epigenomics 2
Session: Genomics/Epigenomics 2
395 - Elucidating Distinct Genetic Signatures in Rhabdomyosarcoma: Potential for Personalized Treatment in Embryonal and Alveolar Subtypes
Friday, April 25, 2025
5:30pm - 7:45pm HST
Publication Number: 395.4910
Shannon Hubany, University of Central Florida College of Medicine, Orlando, FL, United States; Sarah Voskamp, University of Central Florida College of Medicine, Orlando, FL, United States; Allison Dell'Orto, Touro College of Osteopathic Medicine, East Islip, NY, United States; Morgan Engel, University of Central Florida College of Medicine, Orlando, FL, United States; Robert Akins, NemoursAlfred I. duPont Hospital for Children, Wilmington, DE, United States; Jennifer S. Nelson, University of Central Florida College of Medicine, Orlando, FL, United States

Shannon S. Hubany, BS (she/her/hers)
Medical Student
University of Central Florida College of Medicine
Orlando, Florida, United States
Presenting Author(s)
Background: Rhabdomyosarcoma (RMS) is a common soft tissue sarcoma in children and adolescents. The two major histologic subtypes include embryonal (ERMS) and alveolar (ARMS). Despite advancements in multimodal therapy, improving survival for intermediate and high-risk RMS has been unsuccessful.
Objective: In this study we identified distinct genetic signatures associated with ARMS and ERMS and highlighted potential therapeutic targets for individualized treatment according to histologic subtype.
Design/Methods: The Search Tag Analyze Resource for NCBI’s Gene Expression Omnibus was used to identify 36 ERMS, 30 ARMS, and 21 control skeletal muscle samples from microarray data. Over 18,000 genes were extracted using STARGEO meta-analysis. Data were filtered to genes with a statistically significant (p < .05) difference and an absolute experimental log ratio > 0.05. We found 389 ERMS (179 down- and 210 up-regulated) and 341 ARMS (210 down- and 131 up-regulated) genes, which were analyzed using Qiagen's Ingenuity Pathway Analysis (IPA).
Results: ERMS and ARMS showed starkly different genetic signatures with only 218 differentially expressed genes found in both. The only overlap in the top 10 up and down regulated gene candidates was PPP1R12B, which encodes myosin phosphatase subunits MYTP2 and M20. Upregulation of TBX3 was observed in ERMS, which has been previously described in RMS and regulates developmental processes via encoded transcription factors. MEST, a paternally imprinted gene involved in development of mesodermal tissues, was upregulated in ARMS. Top upstream regulators were SDC2 (z-score 1.103), TGFB1 (0.084), and ARID1A (-1.599) for ARMS and ELF3-AS1 (-3.138), DNMT3A (0.577), and ARID3A (2.828) for ERMS. ARID1A contributes to the increased expression of MEST in ARMS. Top canonical pathways in common between ERMS and ARMS include chromatin organization (activated), myelination signaling pathway (inhibited), and non-homologous end joining (activated). Variation occurs with RAR activation pathway (activated in ARMS, inhibited in ERMS) and deubiquitination (activated in ERMS, inhibited in ARMS).
Conclusion(s): Differentially regulated genes in ERMS and ARMS contribute to transcriptional and protein regulation, inflammatory signaling, and skeletal muscle functioning while differentially regulated genes in ARMS are additionally involved in embryonic development of mesodermal tissue, such as skeletal muscle. Tailoring therapies based on distinct genetic signatures may improve outcomes, particularly in high and intermediate-risk patients where current treatments have limited success.
Gene Candidates
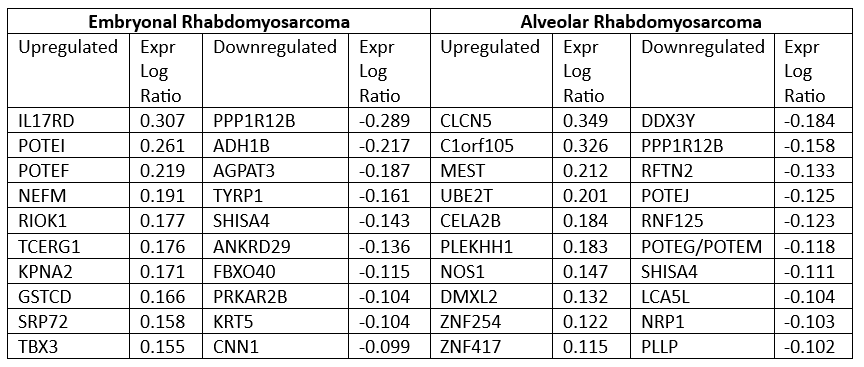 Top 10 up and down regulated genes, ranked according to experimental log ratio, for embryonal rhabdomyosarcoma (left) and alveolar rhabdomyosarcoma (right). Experimental log ratio indicates the magnitude of change on a log base 2 scale between case and control.
Top 10 up and down regulated genes, ranked according to experimental log ratio, for embryonal rhabdomyosarcoma (left) and alveolar rhabdomyosarcoma (right). Experimental log ratio indicates the magnitude of change on a log base 2 scale between case and control.Canonical Pathways ERMS vs ARMS
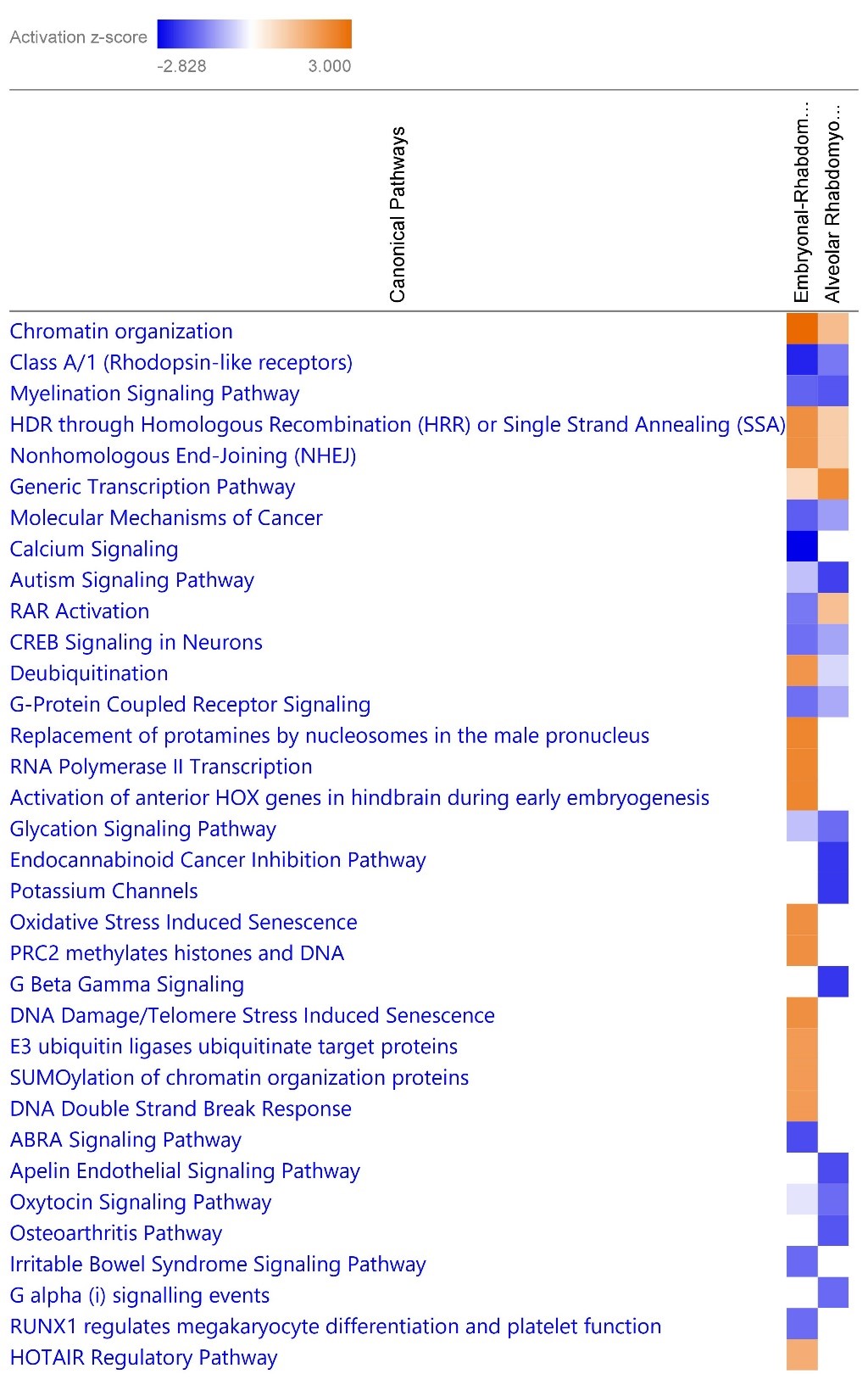 Comparison between the canonical pathways with predicted differential expression in ERMS and ARMS. Blue indicates predicted inhibition and orange indicates predicted activation. Canonical pathways with z-score >1.5 or <-1.5 are included in the heatmap.
Comparison between the canonical pathways with predicted differential expression in ERMS and ARMS. Blue indicates predicted inhibition and orange indicates predicted activation. Canonical pathways with z-score >1.5 or <-1.5 are included in the heatmap. ARID1A, Upstream Regulator of ARMS
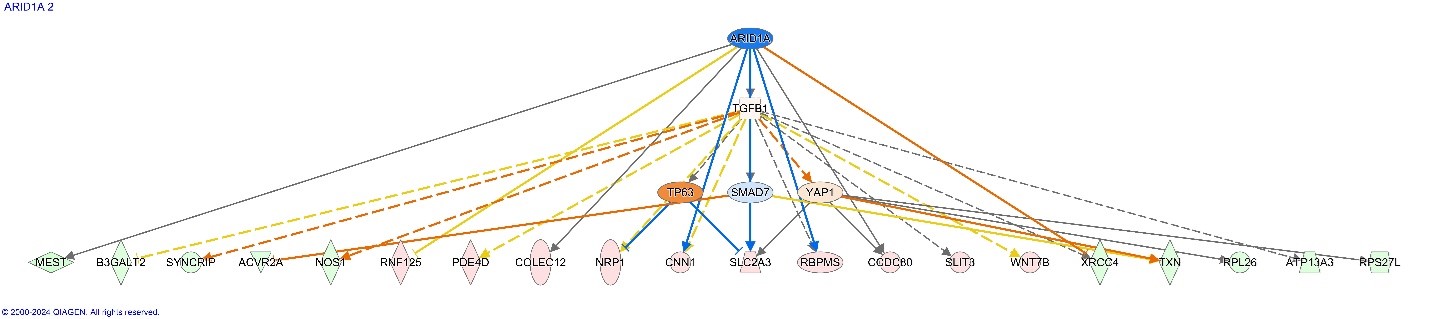 This figure depicts the top upstream regulator, ARID1A, for Alveolar Rhabdomyosarcoma with the downstream effects predicted to ensue from inhibition of ARID1A.
This figure depicts the top upstream regulator, ARID1A, for Alveolar Rhabdomyosarcoma with the downstream effects predicted to ensue from inhibition of ARID1A. 
