Global Neonatal & Children's Health 3
Session: Global Neonatal & Children's Health 3
761 - Metabolomic profiling of biomarkers associated with preterm birth from infants in low resource settings using machine learning
Sunday, April 27, 2025
8:30am - 10:45am HST
Publication Number: 761.5919
Victoria Ward, Stanford University, San Francisco, CA, United States; Ivana Maric, Stanford University School of Medicine, Stanford, CA, United States; Khusbu Adhikari, Stanford University School of Medicine, Stanford, CA, United States; HildaAngela Mujuru, University of Zimbabwe, Harare, Harare, Zimbabwe; Gwendoline Lilly. Chimhini, University of Zimbabwe Faculty of Medicine and Health Sciences, HARARE, Harare, Zimbabwe; Ronald J. Wong, Stanford University School of Medicine, Stanford, CA, United States; Gary Shaw, Stanford University School of Medicine, stanford, CA, United States; Steven Hawken, Ottawa Hospital Research Institute and University of Ottawa, Ottawa, ON, Canada; David K. Stevenson, Stanford University School of Medicine, Stanford, CA, United States; Gary Darmstadt, Department of Pediatrics, Stanford University School of Medicine, Palo Alto, CA, United States

Victoria Ward, MD
Clinical Associate Professor
Stanford University
San Francisco, California, United States
Presenting Author(s)
Background: Preterm birth is the leading cause of neonatal morbidity and mortality worldwide, yet mechanisms for early identification and preventive interventions are lacking. We analyzed machine learning (ML) methods to establish metabolomic profiles of preterm birth from neonatal heel and cord blood.
Objective: The objective was determine if metabolomic biomarkers could be used to accurately predict prematurity.
Design/Methods: Biosamples were collected from infants in three low-resource settings: Bangladesh, Kenya and Zimbabwe. 3,547 infant heel-prick and 2,421 cord blood samples were analyzed for 94 metabolites. Four ML models were used to identify metabolites associated with prematurity. The accuracy, in terms of area under receiver operating characteristic curve (AUROC), was evaluated by cross-validation and by true validation: models trained on multiple cohorts were validated by testing on the remaining cohort.
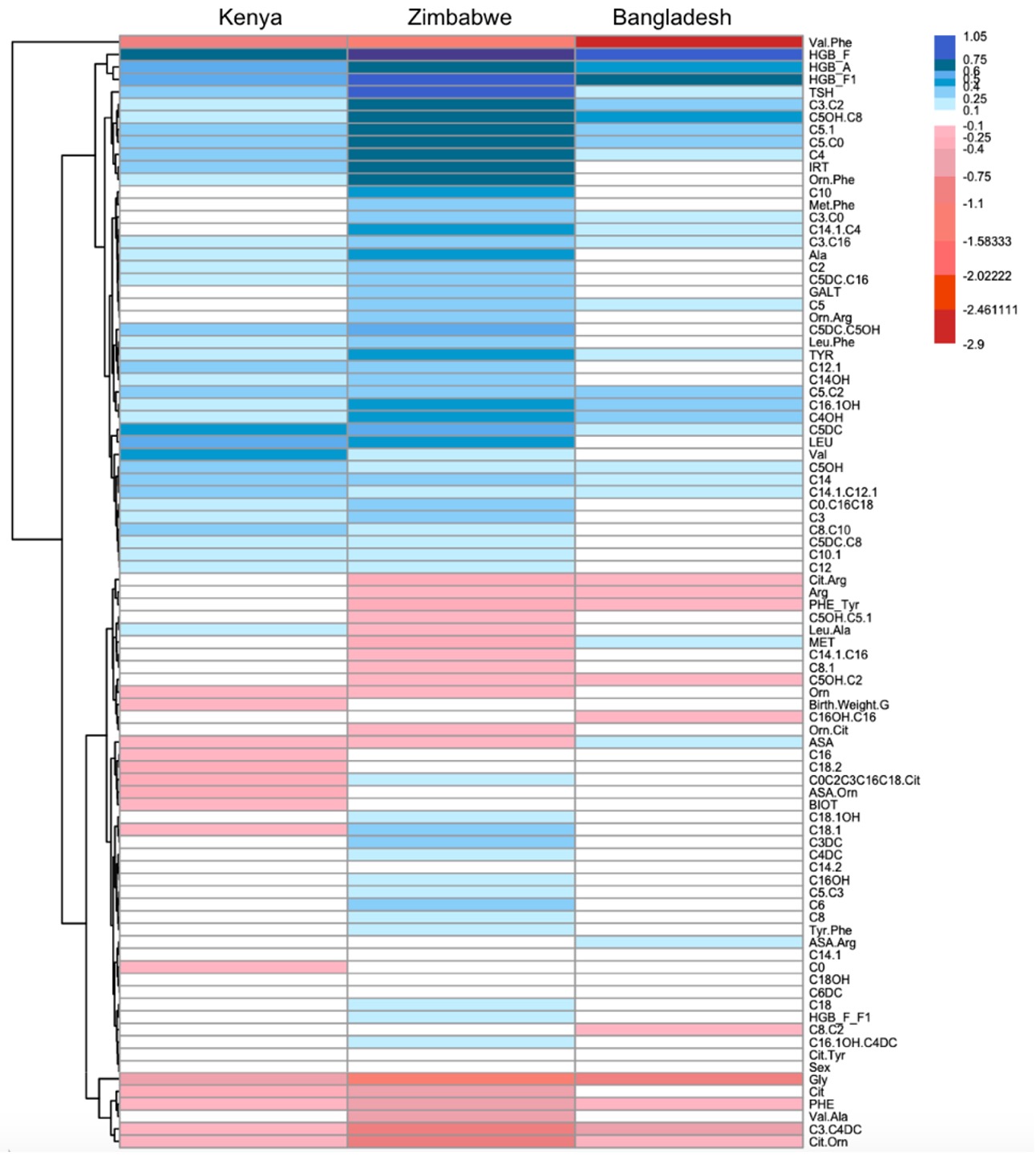
Results: The prevalence of prematurity ranged from 8.8% (Kenya) to 26.2% (Bangladesh). Modest to high accuracy of profiling was achieved across models. Models performed better when tested on the Zimbabwe cohort. Model 1 (Lasso trained on one cohort) had the best performance when trained on data from Kenya and tested on data from Zimbabwe for both heel-prick and cord blood (AUROC (95% CI) 0.816 (0.809, 0.823) and 0.821 (0.817, 0.825), respectively). Model 2 (Lasso trained on multiple concatenated cohorts) and Model 3 (Invariant Causal Prediction trained across cohorts) outperformed Model 1. Model 3 provided similar performance as Model 2 with a smaller number of biomarkers. Model 4 (Stabl) performance varied depending on the training and test cohorts. Biomarkers identified by Models 2, 3, and 4 overlapped (see Table). Stabl biomarkers were a subset of Lasso biomarkers. When cross-validated on the Zimbabwe cohort, AUROCs were 0.824 (Model 2), 0.824 (Model 3), and 0.820 (Model 4). A heatmap was created to summarize regression coefficients for each metabolite across the Zimbabwe, Kenya, and Bangladesh cohorts for heel-prick blood (see Figure).
Conclusion(s): These results create the opportunity for further investigations of metabolomic changes associated with preterm birth. Further, they provide an important step toward developing simple diagnostic tests to identify preterm infants and guiding efficient delivery of high-quality care for those at risk.
Table: Biomarkers Associated with Preterm birth, Kenya and Bangladesh cohorts
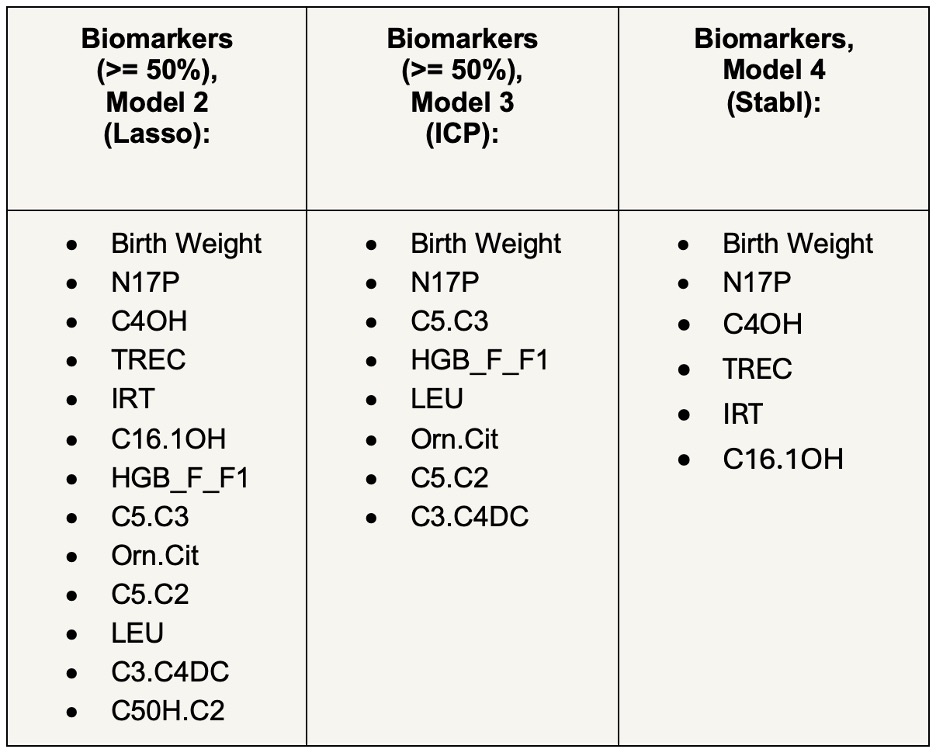
Figure: Differences and Similarities between Regression Coefficients for Biomarkers Between Cohorts