Genomics/Epigenomics 2
Session: Genomics/Epigenomics 2
393 - Deciphering the Therapeutic Effect of Dynamin-2 Reduction in SPEG-Related Myopathy: Perspectives from Multi-Omics Analysis
Friday, April 25, 2025
5:30pm - 7:45pm HST
Publication Number: 393.4253
Qifei Li, University of Miami Miller School of Medicine and Holtz Children's Hospital, Miami, FL, United States; Shiyu Luo, University of Miami Leonard M. Miller School of Medicine, Miami, FL, United States; Jasmine Lin, Boston Children's Hospital, Boston, MA, United States; Haoyue Sheng, University of Miami, Miami, FL, United States; Madesh Ramesh, University of Miami Leonard M. Miller School of Medicine, Miami, FL, United States; Klaus Schmitz-Abe, University of Miami Leonard M. Miller School of Medicine, Miami, FL, United States; Pankaj B.. Agrawal, University of Miami Leonard M. Miller School of Medicine, Miami, FL, United States

Qifei Li, PhD (he/him/his)
Assistant Professor
University of Miami Miller School of Medicine and Holtz Children's Hospital
Miami, Florida, United States
Presenting Author(s)
Background: Striated preferentially expressed protein kinase (SPEG), which belongs to the myosin light chain kinase protein family, is mutated in centronuclear myopathy (CNM) and/or dilated cardiomyopathy. Currently, no therapeutic options are available for this disorder. Dynamin-2 (DNM2) is a member of the large GTPase family, and reducing its levels has been shown to revert muscle phenotypes in mouse models of CNM due to MTM1, DNM2 and BIN1 mutations. We have previously shown that a lack of SPEG results in an increase in DNM2 and that DNM2 reduction by crossing with Dnm2-haploinsufficient mice improves the skeletal muscle function of SPEG-deficient (Speg-CKO) mice. Nevertheless, the molecular mechanisms underlying the amelioration of SPEG-associated myopathy by DNM2 reduction remain elusive.
Objective: We employed multi-omics techniques, including transcriptomic, proteomic, and phosphoproteomic analyses, to elucidate the molecular mechanisms by which DNM2 reduction mitigates the myopathy phenotype in SPEG-related myopathy and to delineate the implicated proteins and pathways.
Design/Methods: Skeletal muscles from adult control, Speg-CKO, and Speg-CKO with DNM2 haploinsufficient (Speg-CKO/DNM2+/-) mice were used for bulk RNA sequencing to profile transcriptomics and tandem mass tags (TMT) mass spectrometry to profile quantitative proteomics and phosphoproteomics.
Results: Transcriptomic analyses indicate that reducing DNM2 levels rescues transcripts associated with phagocytosis and muscle contraction in Speg-CKO skeletal muscles, though it does not restore transcripts related to ECM organization or fatty acid metabolic processes. Additionally, quantitative proteomic analyses show that reducing DNM2 reduction does not restore proteins involved in stabilizing excitation-contraction coupling complexes, such as CMYA5 and FSD2, in SPEG-deficient skeletal muscles. However, this reduction improves proteins associated with T-tubule organization and calcium regulation by the sarcoplasmic reticulum into the cytosol. Phosphoproteomic analyses reveal that 140 differentially expressed phosphosites (DEpPs) are shared between Speg-CKO and Speg-CKO/DNM2+/- muscles compared to control muscle. Additionally, when comparing Speg-CKO to Speg-CKO/DNM2+/- muscles, 30 DEpPs associated with calmodulin binding, actin filament interaction, and protein kinase binding were identified.
Conclusion(s): This work highlights the advantages of using multi-omics techniques to conduct a comprehensive analysis of both pathological and therapeutic changes in rare diseases.
Figure 1. Dynamin-2 reduction rescues the skeletal myopathy of a SPEG-deficient mouse model.
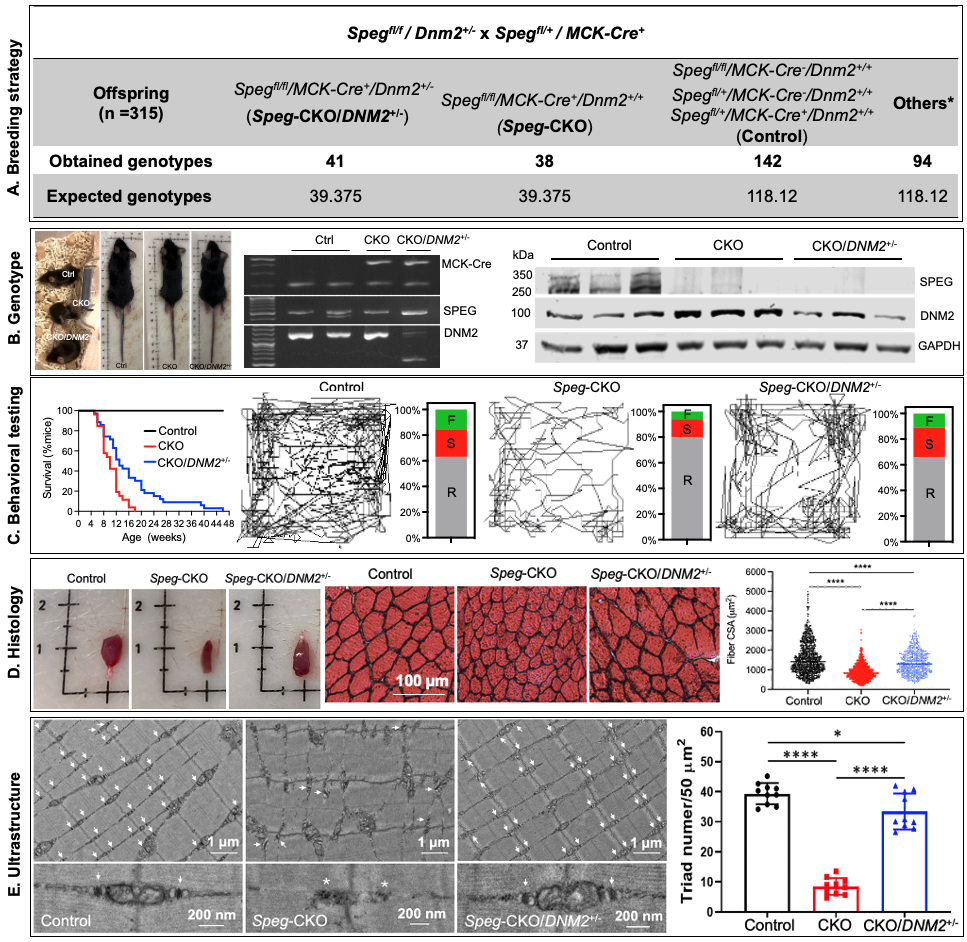 (A) Breeding strategy and outcome for Speg-CKO mice with DNM2 haploinsufficiency (Spegfl/fl/MCK-Cre+/Dnm2+/-: Speg-CKO/DNM2+/-) with expected mice and obtained at 21 days after birth in this study. (B) Left: Representative images of control, Speg-CKO, and Speg-CKO/DNM2+/- mice at 3 months of age and agarose gel analysis of DNA isolated from tails of mice showing the presence or absence of MCK-Cre (top), floxed Speg (middle), and DNM2 (bottom). MCK-Cre+ mice displayed bands for the transgene (~450 bp) and internal positive control (200 bp) while the floxed Speg allele is 485 bp in size versus 422 bp for the WT. DNM2-heterozygous mice displayed 2 bands at 553 bp and 1432 bp versus 1 band at 1432 bp for the WT. Ctrl, control; CKO, Speg-CKO; CKO/DNM2+/-, Speg-CKO/DNM2+/-. Right: Immunoblot analysis of SPEG, DNM2, and GAPDH in skeletal muscles. (C) Left: Life span of Speg-CKO/DNM2+/-, Speg-CKO, and control mice was monitored until 48 weeks of age. Data were represented as percentage survival (control, n = 35; CKO, n = 26; CKO/DNM2+/-, n = 35). Right: Representative mouse activity maps from each genotype of mice and the corresponding activity type distribution graphs at 2 months of age. A clear improvement in ability to explore the arena is observed in Speg-CKO/DNM2+/- mice as compared with Speg-CKO mice. R, resting time; S, slow movement; F, fast movement. (D) Representative TA muscle and H&E staining images at 3 months of age. The mean CSA of Speg-CKO/DNM2+/- TA muscles is significantly larger than that of Speg-CKO (****P < 0.0001, over 500 fibers were analyzed from each group; 1-way ANOVA with Tukey’s post hoc test). (E) Electron micrographs in quadriceps specimens obtained from Speg-CKO/DNM2+/-, Speg-CKO, and control mice at 3 months of age. The upper panel shows an overall organization of muscle structure, and the lower panel shows an enlarged view of triad ultrastructure (white arrows) from each group. Abnormal and fewer triads (white asterisk) are noted in Speg-CKO mice. The number of triads per 50 μm2 was significantly decreased in Speg-CKO mice compared with control mice. However, the triad number in Speg-CKO/DNM2+/- mice was remarkably increased compared with Speg-CKO mice. Each dot represents a randomly selected field to count the triad number; *P ˂ 0.05; ****P < 0.0001; n = 3 per genotype.
(A) Breeding strategy and outcome for Speg-CKO mice with DNM2 haploinsufficiency (Spegfl/fl/MCK-Cre+/Dnm2+/-: Speg-CKO/DNM2+/-) with expected mice and obtained at 21 days after birth in this study. (B) Left: Representative images of control, Speg-CKO, and Speg-CKO/DNM2+/- mice at 3 months of age and agarose gel analysis of DNA isolated from tails of mice showing the presence or absence of MCK-Cre (top), floxed Speg (middle), and DNM2 (bottom). MCK-Cre+ mice displayed bands for the transgene (~450 bp) and internal positive control (200 bp) while the floxed Speg allele is 485 bp in size versus 422 bp for the WT. DNM2-heterozygous mice displayed 2 bands at 553 bp and 1432 bp versus 1 band at 1432 bp for the WT. Ctrl, control; CKO, Speg-CKO; CKO/DNM2+/-, Speg-CKO/DNM2+/-. Right: Immunoblot analysis of SPEG, DNM2, and GAPDH in skeletal muscles. (C) Left: Life span of Speg-CKO/DNM2+/-, Speg-CKO, and control mice was monitored until 48 weeks of age. Data were represented as percentage survival (control, n = 35; CKO, n = 26; CKO/DNM2+/-, n = 35). Right: Representative mouse activity maps from each genotype of mice and the corresponding activity type distribution graphs at 2 months of age. A clear improvement in ability to explore the arena is observed in Speg-CKO/DNM2+/- mice as compared with Speg-CKO mice. R, resting time; S, slow movement; F, fast movement. (D) Representative TA muscle and H&E staining images at 3 months of age. The mean CSA of Speg-CKO/DNM2+/- TA muscles is significantly larger than that of Speg-CKO (****P < 0.0001, over 500 fibers were analyzed from each group; 1-way ANOVA with Tukey’s post hoc test). (E) Electron micrographs in quadriceps specimens obtained from Speg-CKO/DNM2+/-, Speg-CKO, and control mice at 3 months of age. The upper panel shows an overall organization of muscle structure, and the lower panel shows an enlarged view of triad ultrastructure (white arrows) from each group. Abnormal and fewer triads (white asterisk) are noted in Speg-CKO mice. The number of triads per 50 μm2 was significantly decreased in Speg-CKO mice compared with control mice. However, the triad number in Speg-CKO/DNM2+/- mice was remarkably increased compared with Speg-CKO mice. Each dot represents a randomly selected field to count the triad number; *P ˂ 0.05; ****P < 0.0001; n = 3 per genotype.Figure 2. Multi-omics insights into DNM2 reduction-mediated myopathy alleviation in SPEG-deficient mice.
Fig. 2.pdf(A) Transcriptome profiling in the skeletal muscle of Speg-CKO mice with DNM2 haploinsufficiency. (B) Overview of quantitative proteomics and phosphoproteomics profiling in the skeletal muscle of Speg-CKO mice. (C) Proteome profiling in the skeletal muscle of Speg-CKO mice with DNM2 haploinsufficiency. Volcano plots represent the differentially expressed proteins (DEPs). Upregulated proteins are in red, and downregulated proteins are in blue (Padj <0.1 and a fold change beyond ±1.5). Heat map and gene ontology (GO) analysis of the DEPs.
Figure 3. Phosphoproteomics analysis in the skeletal muscle of Speg-CKO mice with DNM2 haploinsufficiency.
Fig 3.pdf(A) Volcano plots represent the differentially expressed phosphosites (DEpPs). Upregulated DEpPs are in red, and downregulated DEpPs are in blue (Padj <0.1 and a fold change beyond ±1.5). (B) Venn plot of DEpPs analysis among three groups and the corresponding Heat map and gene ontology (GO) analysis of the DEpPs.

